
The rhinovirus is the most common viral infectious agent in humans and is the predominant cause of the common cold. Rhinovirus infection proliferates in temperatures of 33–35 °C (91–95 °F), the temperatures found in the nose. Rhinoviruses belong to the genus Enterovirus in the family Picornaviridae.
Coxsackie B4 virus are enteroviruses that belong to the Picornaviridae family. These viruses can be found worldwide. They are positive-sense, single-stranded, non-enveloped RNA viruses with icosahedral geometry. Coxsackieviruses have two groups, A and B, each associated with different diseases. Coxsackievirus group A is known for causing hand-foot-and-mouth diseases while Group B, which contains six serotypes, can cause a varying range of symptoms like gastrointestinal distress myocarditis. Coxsackievirus B4 has a cell tropism for natural killer cells and pancreatic islet cells. Infection can lead to beta cell apoptosis which increases the risk of insulitis.

Poliovirus, the causative agent of polio, is a serotype of the species Enterovirus C, in the family of Picornaviridae. There are three poliovirus serotypes: types 1, 2, and 3.

Picornaviruses are a group of related nonenveloped RNA viruses which infect vertebrates including fish, mammals, and birds. They are viruses that represent a large family of small, positive-sense, single-stranded RNA viruses with a 30 nm icosahedral capsid. The viruses in this family can cause a range of diseases including the common cold, poliomyelitis, meningitis, hepatitis, and paralysis.

Foot-and-mouth disease virus (FMDV) is the pathogen that causes foot-and-mouth disease. It is a picornavirus, the prototypical member of the genus Aphthovirus. The disease, which causes vesicles (blisters) in the mouth and feet of cattle, pigs, sheep, goats, and other cloven-hoofed animals is highly infectious and a major plague of animal farming.

Rubella virus (RuV) is the pathogenic agent of the disease rubella, transmitted only between humans via the respiratory route, and is the main cause of congenital rubella syndrome when infection occurs during the first weeks of pregnancy.
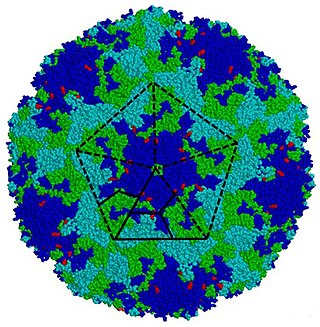
Enterovirus is a genus of positive-sense single-stranded RNA viruses associated with several human and mammalian diseases. Enteroviruses are named by their transmission-route through the intestine.

Orbivirus is a genus of double-stranded RNA viruses in the family Reoviridae and subfamily Sedoreovirinae. Unlike other reoviruses, orbiviruses are arboviruses. They can infect and replicate within a wide range of arthropod and vertebrate hosts. Orbiviruses are named after their characteristic doughnut-shaped capsomers.
Parechovirus B, formerly called the Ljungan virus, was first discovered in the mid-1990s after being isolated from a bank vole near the Ljungan river in Medelpad county, Sweden. It has since been established that Parechovirus B, which is also found in several places in Europe and America, causes serious illness in wild as well as laboratory animals. Several scientific articles have recently reported findings indicating that Parechovirus B is associated with malformations, intrauterine fetal death, and sudden infant death syndrome in humans. In addition, studies are being conducted worldwide to investigate the possible connection of the virus to diabetes, neurological and other illnesses in humans.
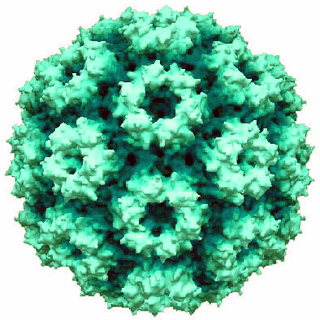
Cowpea chlorotic mottle virus, known by the abbreviation CCMV, is a virus that specifically infects the cowpea plant, or black-eyed pea. The leaves of infected plants develop yellow spots, hence the name "chlorotic". Similar to its "brother" virus, Cowpea mosaic virus (CPMV), CCMV is produced in high yield in plants. In the natural host, viral particles can be produced at 1–2 mg per gram of infected leaf tissue. Belonging to the bromovirus genus, cowpea chlorotic mottle virus (CCMV) is a small spherical plant virus. Other members of this genus include the brome mosaic virus (BMV) and the broad bean mottle virus (BBMV).
Erbovirus is a genus of viruses in the order Picornavirales, in the family Picornaviridae. Horses serve as natural hosts. There is only one species in this genus: Erbovirus A. Diseases associated with this genus include: upper respiratory tract disease with viremia and fecal shedding. Viruses belonging to the genus Erbovirus have been isolated in horses with acute upper febrile respiratory disease. The structure of the Erbovirus virion is icosahedral, having a diameter of 27–30 nm.

This family represents the internal ribosome entry site (IRES) of the hepatitis A virus. HAV IRES is a 450 nucleotide long sequence located in the 735 nt long 5’ UTR of Hepatitis A viral RNA genome. IRES elements allow cap and end-independent translation of mRNA in the host cell. The IRES achieves this by mediating the internal initiation of translation by recruiting a ribosomal 40S pre-initiation complex directly to the initiation codon and eliminates the requirement for eukaryotic initiation factor, eIF4F.

Enterovirus E is a picornavirus of the genus Enterovirus. The virus may also be referred to as enteric cytopathic bovine orphan virus (ECBO). It is endemic in cattle populations worldwide, and although normally fairly nonpathogenic, it can cause reproductive, respiratory, or enteric disease – particularly when the animal is concurrently infected with another pathogen.
Chronic Mycoplasma pneumonia and Chlamydia pneumonia infections are associated with the onset and exacerbation of asthma. These microbial infections result in chronic lower airway inflammation, impaired mucociliary clearance, an increase in mucous production and eventually asthma. Furthermore, children who experience severe viral respiratory infections early in life have a high possibility of having asthma later in their childhood. These viral respiratory infections are mostly caused by respiratory syncytial virus (RSV) and human rhinovirus (HRV). Although RSV infections increase the risk of asthma in early childhood, the association between asthma and RSV decreases with increasing age. HRV on the other hand is an important cause of bronchiolitis and is strongly associated with asthma development. In children and adults with established asthma, viral upper respiratory tract infections (URIs), especially HRVs infections, can produce acute exacerbations of asthma. Thus, Chlamydia pneumoniae, Mycoplasma pneumoniae and human rhinoviruses are microbes that play a major role in non-atopic asthma.

Picornain 3C is a protease found in picornaviruses, which cleaves peptide bonds of non-terminal sequences. Picornain 3C’s endopeptidase activity is primarily responsible for the catalytic process of selectively cleaving Gln-Gly bonds in the polyprotein of poliovirus and with substitution of Glu for Gln, and Ser or Thr for Gly in other picornaviruses. Picornain 3C are cysteine proteases related by amino acid sequence to trypsin-like serine proteases. Picornain 3C is encoded by enteroviruses, rhinoviruses, aphtoviruses and cardioviruses. These genera of picoviruses cause a wide range of infections in humans and mammals.
Mastadenovirus is a genus of viruses in the family Adenoviridae. Humans and other mammals serve as natural hosts. There are 51 species in this genus. The genus as a whole includes many very common causes of human infection, estimated to be responsible for 2 to 5% of all respiratory infections, as well as gastrointestinal and eye infections. Symptoms are usually mild.

Avibirnavirus is a genus of viruses in family Birnaviridae. There is a single species in this genus: Infectious bursal disease virus, which infects chickens and other fowl. It causes severe inflammation of the bursa of Fabricius, and causes considerable morbidity and mortality.
Triatoma virus (TrV) is a virus belonging to the insect virus family Dicistroviridae. Within this family, there are currently 3 genera and 15 species of virus. Triatoma virus belongs to the genus Cripavirus. It is non-enveloped and its genetic material is positive-sense, single-stranded RNA. The natural hosts of triatoma virus are invertebrates. TrV is a known pathogen to Triatoma infestans, the major vector of Chagas disease in Argentina which makes triatoma virus a major candidate for biological vector control as opposed to chemical insecticides. Triatoma virus was first discovered in 1984 when a survey of pathogens of triatomes was conducted in the hopes of finding potential biological control methods for T. infestans.
Black beetle virus (BBV) is a virus that was initially discovered in the North Island of New Zealand in Helensville in dead New Zealand black beetles in 1975.

Monodnaviria is a realm of viruses that includes all single-stranded DNA viruses that encode an endonuclease of the HUH superfamily that initiates rolling circle replication of the circular viral genome. Viruses descended from such viruses are also included in the realm, including certain linear single-stranded DNA (ssDNA) viruses and circular double-stranded DNA (dsDNA) viruses. These atypical members typically replicate through means other than rolling circle replication.















