See also
Genetics
Y-DNA O subclades
Y-DNA backbone tree
| |||
Footnotes
| |||
| Haplogroup O-M176 | |
|---|---|
| Possible time of origin | 31,108 (95% CI 22,844 <-> 34,893) years before present [1] 29,190 years before present [2] 28,200 [95% CI 26,000 <-> 30,400] years before present [3] |
| Coalescence age | 25,660 years before present [2] 25,800 [95% CI 23,400 <-> 28,400] years before present (YFull [3] [4] ) |
| Possible place of origin | the Korean Peninsula or a nearby part of northeastern Asia [5] |
| Ancestor | O-P31 |
| Defining mutations | M176/SRY465, P49, 022454[ citation needed ] |
| Highest frequencies | Japanese, Koreans, Ryukyuans, Manchus
|
Haplogroup O-M176 (aka O-SRY465) or O1b2 is a human Y-chromosome DNA haplogroup. It is best known for its part in the settlement of Korea and Japan. It is a descendant of Haplogroup O-P31, and it has been estimated to share a most recent common ancestor with its nearest outgroup, Haplogroup O-K18, approximately 31,108 (95% CI 22,844 <-> 34,893) years before present, [1] approximately 29,190 years before present, [2] or approximately 28,200 (95% CI 26,000 <-> 30,400) years before present. [3]
Haplogroup O-M176 is found mainly in the northernmost parts of East Asia, from the Uriankhai and Zakhchin peoples of western Mongolia ( Katoh et al. 2005 ) to the Japanese of Japan, though it also has been detected sporadically in the Buryats ( Jin et al. 2003 ). It has been detected with moderate frequencies in Udegeys ( Jin, Kim & Kim 2010 ) of southern Siberia, rarely among populations of Southeast Asia including Indonesia (Hammer et al. 2006 and Jin et al. 2003), the Philippines ( Jin et al. 2003 ), Thailand ( Jin et al. 2003 ), and Vietnam (Hammer et al. 2006 and Jin et al. 2003), and Micronesians ( Hammer et al. 2006 ). This haplogroup is found with its highest frequency and diversity values among modern populations of Japan and Korea and is rare in most populations in China. Among Han Chinese, it has been detected in some samples of Han Chinese from Beijing (1/51, Jin et al. 2003 and Kim et al. 2011), [5] Xi'an (1/34, Kim et al. 2011), [5] one Han Chinese in Henan, [19] Han Chinese in Taiwan (2/352 = 0.57%, including one of 34 Hakka people and one of 258 miscellaneous Han volunteers), [20] Han Chinese from East China sampled from the infertility clinic at the Affiliated Hospitals of Nanjing Medical University at Jiangsu (6/1147 = 0.52%, Lu et al. 2009), Wuhan (1/160), [21] and South China outside of Jiangsu, Anhui, Zhejiang, and Shanghai (1/65). [22] Among ethnic minorities in China, haplogroup O-M176 has been detected with high frequency in samples of Koreans in China (Xue et al. 2006 and Katoh et al. 2005) and with low frequency among Manchus [23] (Xue et al. 2006, Katoh et al. 2005, and Karafet et al. 2001), Hezhe people, [12] Daurs, [12] Evenks, [11] Sibes ( Xue et al. 2006 ), Kham Tibetans, [24] and Hui. [25] In a study of various populations of Hunan, O1b2-M176 was found in 0.55% (5/903) of all samples; specifically, this haplogroup was observed in 3.0% (1/33) of a sample of Iu Mien from Hunan, 1.9% (2/103) of a sample of Gàn Chinese from Hunan, 1.4% (1/71) of a sample of Kam from Hunan, and 1.1% (1/95) of a sample of Xong Miao from Hunan. [26] In a study published in July 2020, Y-DNA belonging to haplogroup O1b2-M176 was observed in 1.31% (4/305) of a sample of Han Chinese from Zibo, Shandong and in 1.06% (6/565) of a sample of Han Chinese from Zhaotong, Yunnan. [27]
Mitsuru Sakitani suggests that haplogroup O1b2, which is common in today Koreans, Japanese and Manchu, are one of the carriers of Liao civilization or Yangtze civilization. As the Liao civilization and the Yangtze civilization declined several tribes crossed westward and northerly, to the Korean Peninsula and the Japanese archipelago. However, Mitsuru Sakitani said that Currently, very little o1b2 are detected in the Yangtze River region, there are many problems in the theory that originate from the Yangtze River area. [28] [29] [30] Another study calls the haplogroup O1b1 as the major Austroasiatic paternal lineage and the haplogroup O1b2 (of Koreans and Japanese) as the "para-Austroasiatic" paternal lineage. [31]
Y-DNA that belongs to O-M176(xK10, F3356) has been found in an individual from Hiroshima, [4] an individual from Fukushima, [4] an individual from Beijing, [4] and 1% (7/706) of a sample of males collected in Seoul and Daejeon. [32]
O-M176(x47z) has been found in approximately 9.2% of Japanese males (ranging from 3.5% in the JPT sample from Tokyo [33] to 13.1% in a sample from Shizuoka [34] ) and in approximately 8.3% of Ryukyuan males (ranging from 5.3% in a sample from Miyako [8] to 11.1% in a sample from Okinawa [34] ). [6]
The majority of extant members of O-M176 belong to the subclade O-K10 (aka O-F3356 aka O-F1204). O-K10 (TMRCA 8,070 ybp according to TheYtree, [35] 7,900 [95% CI 5,624 <-> 9,449] ybp according to Karmin et al. 2022, [1] 7,457 (99% CI 9,434 - 5,789) years before present according to FamilyTreeDNA, [36] 7,000 [95% CI 8,000 <-> 6,000] ybp according to YFull, [4] or 6,970 years according to 23mofang [2] ) subsumes the prolific subclades O-47z, which occurs with especially high frequency in Japan, and O-L682, which occurs with especially high frequency in Korea, in addition to the relatively rare subclades O-CTS10687, which has been found in Japan, Korea, and China, and O-K3, which has been found among Han Chinese mostly in South Central China. O-L682 and O-K3 are linked by 18 SNPs that define the O-K4 clade, and thus their members are more closely related to one another by paternal lineage than any of them is related to any member of O-47z or O-CTS10687.
O-F3356(x47z, L682) has been found in 2% (14/706) of a sample of Koreans collected in Seoul and Daejeon, South Korea. [32] However, the status of these individuals' Y-DNA in regard to K4, K3, CTS10687, and phylogenetically equivalent SNPs has not been published.
O-CTS10687 has been found in 1.8% (1/56) of the JPT sample of Japanese from Tokyo, Japan. [37] [4]
| Haplogroup O-47z | |
|---|---|
| Possible time of origin | 7,870 [95% CI 5,720–12,630] years ago (Hammer et al. 2006) 7,613 (95% CI 5,309 <-> 9,130) ybp [1] 7,000 [95% CI 6,100 <-> 7,900] ybp [3] |
| Coalescence age | 5,780 ybp [2] 5,600 (95% CI 6,500 <-> 4,700) ybp [4] |
| Possible place of origin | Japanese Archipelago (Hammer et al. 2006) or Korean Peninsula (Jin et al. 2003) |
| Ancestor | O-M176 |
| Defining mutations | 47z |
| Highest frequencies | Japanese, Ryukyuans, Koreans |
O-47z or O-CTS11986 is a subclade of O-K10. It is found with high frequency among the Japanese and Ryukyuan populations of Japan, and with lower frequency among Koreans.
Haplogroup O-47z has been detected in approximately 22% of males who speak a Japonic language, while it has not been found at all among Ainu males whose Y-DNA has been tested in two genetic studies (Tajima et al. 2004, n=16; Hammer et al. 2006, n=4). Based on the STR haplotype diversity within Haplogroup O-47z, it has been estimated in a study published in 2006 that this haplogroup has expanded from a single founder who has lived approximately 3,810 (95% CI 1,640 <–> 7,960) years before present in a model according to which continuous, pure exponential population growth is assumed. [11] In a paper published in 2016, the time to most recent common ancestor of a set of fifteen members of the O-47z clade, all from the JPT (Japanese in Tokyo, Japan) sample of the 1000 Genomes Project, was estimated to be 4,500 years using a relatively slow mutation rate (μ = 0.76 x 10−9 per bp per year as according to Qiaomei Fu et al. 2014) or 3,900 years using a relatively fast mutation rate (μ = 0.888 x 10−9 per bp per year as according to A. Helgason et al. 2015). [33] Haplogroup O-47z also has been found among samples of modern Koreans, though with low frequency in comparison to both the frequency of O-47z in samples of Japanese and the frequency of O-M176(x47z) in samples of Koreans. [5]
Soon-Hee Kim et al. (2011) found haplogroup O-47z (DXYS5Y-Y2) in 8.89% (45/506) of a pool of samples from South Korea. O-47z was found in greatest proportion in the study's sample from the Gyeongsang region (10/84 = 11.9%), which is located in the southeast corner of the Korean Peninsula, and in least proportion in the study's sample from the Seoul-Gyeonggi region (8/110 = 7.3%), which is located on the west coast of the middle of the Korean Peninsula. [38] Haplogroup O-47z also has been observed in a sample of Koreans in China (2/25 = 8.0%). [39]
O-K4 is a subclade of O-K10. It includes at least two subclades, O-L682 and O-K3, which have been estimated to share a most recent common ancestor approximately 6,327 (95% CI 4,575 <-> 7,762) years before present. [1]
| Haplogroup O-F940 | |
|---|---|
| Possible time of origin | 6,327 (95% CI 4,575 <-> 7,762) years before present [1] 6,000 years before present [2] 6,000 (95% CI 7,100 <-> 4,900) ybp [4] |
| Coalescence age | 2,650 years before present [2] |
| Possible place of origin | East Asia (origin) China (MRCA) |
| Ancestor | O-F2868 |
| Defining mutations | CTS12145, F1912, F2206, F2703, F940, K3 |
| Highest frequencies | Chinese [4] [1] 0.12% [2] |
The O-K3 (or O-F940) lineage is a subclade of O-K4 that has been observed to date in three individuals from Hunan, [4] one individual from Jiangxi, [4] and one individual from Henan. [19] The TMRCA of the three individuals from Hunan plus the one individual from Jiangxi has been estimated to be 1,300 (95% CI 800 <-> 2,100) ybp. [4]
| Haplogroup O-L682 | |
|---|---|
| Possible time of origin | 6,327 [95% CI 4,575 <-> 7,762] ybp [1] 5,990 ybp [2] |
| Coalescence age | 4,080 ybp [2] 4,200 (95% CI 4,900 <-> 3,500) ybp [4] |
| Possible place of origin | Korean Peninsula or Manchuria |
| Ancestor | O-M176, O-F3356 |
| Defining mutations | L682 |
| Highest frequencies | Koreans, Japanese, Ryukyuans, Hezhen, Manchus
|
The O-L682 subclade of O-K4 is believed to be related to Native Korean population. One study has found O-L682 Y-DNA in 19% (134/706) of Koreans sampled in Seoul and Daejeon. [32] O-L682 also has been found in Japanese in Tokyo, Okayama, Kōchi, and the US and in Chinese (especially in Jilin, Heilongjiang, and Liaoning, with a greater than average presence also in Beijing, Hebei, Inner Mongolia, Gansu, Shaanxi, Shandong, Tianjin, and Anhui, [2] and with some presence in other areas, such as Shanxi, [4] and among some ethnic minorities, such as Nanai people [4] ). Its descendants appear to have begun rapidly increasing in number at approximately the same time as those of its distant cousin O-47z, perhaps 4,000 years ago. [4]
Prior to 2002, there were in academic literature at least seven naming systems for the Y-Chromosome phylogenetic tree. This led to considerable confusion. In 2002, the major research groups came together and formed the Y-Chromosome Consortium (YCC). They published a joint paper that created a single new tree that all agreed to use. Later, a group of citizen scientists with an interest in population genetics and genetic genealogy formed a working group to create an amateur tree aiming at being above all timely. The table below brings together all of these works at the point of the landmark 2002 YCC Tree. This allows a researcher reviewing older published literature to quickly move between nomenclatures.
| YCC 2002/2008 (Shorthand) | (α) | (β) | (γ) | (δ) | (ε) | (ζ) | (η) | YCC 2002 (Longhand) | YCC 2005 (Longhand) | YCC 2008 (Longhand) | YCC 2010r (Longhand) | ISOGG 2006 | ISOGG 2007 | ISOGG 2008 | ISOGG 2009 | ISOGG 2010 | ISOGG 2011 | ISOGG 2012 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| O-M175 | 26 | VII | 1U | 28 | Eu16 | H9 | I | O* | O | O | O | O | O | O | O | O | O | O |
| O-M119 | 26 | VII | 1U | 32 | Eu16 | H9 | H | O1* | O1a | O1a | O1a | O1a | O1a | O1a | O1a | O1a | O1a | O1a |
| O-M101 | 26 | VII | 1U | 32 | Eu16 | H9 | H | O1a | O1a1 | O1a1a | O1a1a | O1a1 | O1a1 | O1a1a | O1a1a | O1a1a | O1a1a | O1a1a |
| O-M50 | 26 | VII | 1U | 32 | Eu16 | H10 | H | O1b | O1a2 | O1a2 | O1a2 | O1a2 | O1a2 | O1a2 | O1a2 | O1a2 | O1a2 | O1a2 |
| O-P31 | 26 | VII | 1U | 33 | Eu16 | H5 | I | O2* | O2 | O2 | O2 | O2 | O2 | O2 | O2 | O2 | O2 | O2 |
| O-M95 | 26 | VII | 1U | 34 | Eu16 | H11 | G | O2a* | O2a | O2a | O2a | O2a | O2a | O2a | O2a | O2a | O2a1 | O2a1 |
| O-M88 | 26 | VII | 1U | 34 | Eu16 | H12 | G | O2a1 | O2a1 | O2a1 | O2a1 | O2a1 | O2a1 | O2a1 | O2a1 | O2a1 | O2a1a | O2a1a |
| O-SRY465 | 20 | VII | 1U | 35 | Eu16 | H5 | I | O2b* | O2b | O2b | O2b | O2b | O2b | O2b | O2b | O2b | O2b | O2b |
| O-47z | 5 | VII | 1U | 26 | Eu16 | H5 | I | O2b1 | O2b1a | O2b1 | O2b1 | O2b1a | O2b1a | O2b1 | O2b1 | O2b1 | O2b1 | O2b1 |
| O-M122 | 26 | VII | 1U | 29 | Eu16 | H6 | L | O3* | O3 | O3 | O3 | O3 | O3 | O3 | O3 | O3 | O3 | O3 |
| O-M121 | 26 | VII | 1U | 29 | Eu16 | H6 | L | O3a | O3a | O3a1 | O3a1 | O3a1 | O3a1 | O3a1 | O3a1 | O3a1 | O3a1a | O3a1a |
| O-M164 | 26 | VII | 1U | 29 | Eu16 | H6 | L | O3b | O3b | O3a2 | O3a2 | O3a2 | O3a2 | O3a2 | O3a2 | O3a2 | O3a1b | O3a1b |
| O-M159 | 13 | VII | 1U | 31 | Eu16 | H6 | L | O3c | O3c | O3a3a | O3a3a | O3a3 | O3a3 | O3a3a | O3a3a | O3a3a | O3a3a | O3a3a |
| O-M7 | 26 | VII | 1U | 29 | Eu16 | H7 | L | O3d* | O3c | O3a3b | O3a3b | O3a4 | O3a4 | O3a3b | O3a3b | O3a3b | O3a2b | O3a2b |
| O-M113 | 26 | VII | 1U | 29 | Eu16 | H7 | L | O3d1 | O3c1 | O3a3b1 | O3a3b1 | - | O3a4a | O3a3b1 | O3a3b1 | O3a3b1 | O3a2b1 | O3a2b1 |
| O-M134 | 26 | VII | 1U | 30 | Eu16 | H8 | L | O3e* | O3d | O3a3c | O3a3c | O3a5 | O3a5 | O3a3c | O3a3c | O3a3c | O3a2c1 | O3a2c1 |
| O-M117 | 26 | VII | 1U | 30 | Eu16 | H8 | L | O3e1* | O3d1 | O3a3c1 | O3a3c1 | O3a5a | O3a5a | O3a3c1 | O3a3c1 | O3a3c1 | O3a2c1a | O3a2c1a |
| O-M162 | 26 | VII | 1U | 30 | Eu16 | H8 | L | O3e1a | O3d1a | O3a3c1a | O3a3c1a | O3a5a1 | O3a5a1 | O3a3c1a | O3a3c1a | O3a3c1a | O3a2c1a1 | O3a2c1a1 |
The following research teams per their publications were represented in the creation of the YCC Tree.
This phylogenetic tree of haplogroup O subclades is based on the YCC 2008 tree ( Karafet et al. 2008 ) and subsequent published research.

Haplogroup J-M304, also known as J, is a human Y-chromosome DNA haplogroup. It is believed to have evolved in Western Asia. The clade spread from there during the Neolithic, primarily into North Africa, the Horn of Africa, the Socotra Archipelago, the Caucasus, Europe, Western Asia, Central Asia, South Asia, and Southeast Asia.

Haplogroup C is a major Y-chromosome haplogroup, defined by UEPs M130/RPS4Y711, P184, P255, and P260, which are all SNP mutations. It is one of two primary branches of Haplogroup CF alongside Haplogroup F. Haplogroup C is found in ancient populations on every continent except Africa and is the predominant Y-DNA haplogroup among males belonging to many peoples indigenous to East Asia, Central Asia, Siberia, North America and Australia as well as a some populations in Europe, the Levant, and later Japan. The haplogroup is also found with moderate to low frequency among many present-day populations of Southeast Asia, South Asia, and Southwest Asia.
Haplogroup D1 or D-M174 is a subclade of Haplogroup D-CTS3946. This male haplogroup is found primarily in East Asia and the Andaman Islands, though it is also found regularly with low frequency in Central Asia and Mainland Southeast Asia. It's also found as far as Europe and Middle east in lower frequencies.

Haplogroup N (M231) is a Y-chromosome DNA haplogroup defined by the presence of the single-nucleotide polymorphism (SNP) marker M231.
Haplogroup O-M122 is an Eastern Eurasian Y-chromosome haplogroup. The lineage ranges across Southeast Asia and East Asia, where it dominates the paternal lineages with extremely high frequencies. It is also significantly present in Central Asia, especially among the Naiman tribe of Kazakhs.
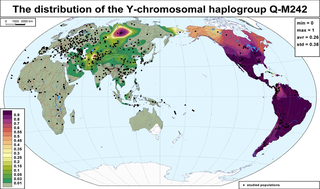
Haplogroup Q or Q-M242 is a Y-chromosome DNA haplogroup. It has one primary subclade, Haplogroup Q1 (L232/S432), which includes numerous subclades that have been sampled and identified in males among modern populations.
Haplogroup R, or R-M207, is a Y-chromosome DNA haplogroup. It is both numerous and widespread amongst modern populations.

Tungusic peoples are an ethno-linguistic group formed by the speakers of Tungusic languages. They are native to Siberia and Northeast Asia.
In human genetics, Haplogroup O-M268, also known as O1b, is a Y-chromosome DNA haplogroup. Haplogroup O-M268 is a primary subclade of haplogroup O-F265, itself a primary descendant branch of Haplogroup O-M175.
In human genetics, Haplogroup O-M119 is a Y-chromosome DNA haplogroup. Haplogroup O-M119 is a descendant branch of haplogroup O-F265 also known as O1a, one of two extant primary subclades of Haplogroup O-M175. The same clade previously has been labeled as O-MSY2.2.

Haplogroup NO, also known as NO-M214 and NO1, is a human Y-chromosome DNA haplogroup. NO is the sole confirmed subclade of Haplogroup K2a1 (K-M2313), which is the sole subclade of Haplogroup K2a (K-M2308). NO is the dominant Y-DNA haplogroup in most parts of eastern and northern Eurasia, including East Asia, Siberia and northern Fennoscandia.

Haplogroup C-M217, also known as C2, is a Y-chromosome DNA haplogroup. It is the most frequently occurring branch of the wider Haplogroup C (M130). It is found mostly in Central Asia, Eastern Siberia and significant frequencies in parts of East Asia and Southeast Asia including some populations in the Caucasus, Middle East, South Asia, East Europe. It is found in a much more widespread areas with a low frequency of less than 2%.
Haplogroup O-K18 also known as O-F2320 and Haplogroup O1b1, is a human Y-chromosome DNA haplogroup. Haplogroup O-K18 is a descendant branch of Haplogroup O-P31. Based on its disjunct distribution, O-K18 can be further divided into south subclade O1b1a1-PK4 and north subclade O1b1a2-CTS4040. O-CTS4040 is widely distributed in East Asia, whereas O-PK4 is more frequent in South China and Southeast Asia. O-PK4 is best known for the high frequency of its O-M95 subclade among populations of Southeast Asia and among speakers of Austroasiatic languages in South Asia.
E-P2, also known as E1b1, is a human Y-chromosome DNA haplogroup. This paternal clade had an ancient presence in the Middle East, and is now primarily distributed in Africa where it's believed to have originated, with lower frequencies in the Middle East and Europe.
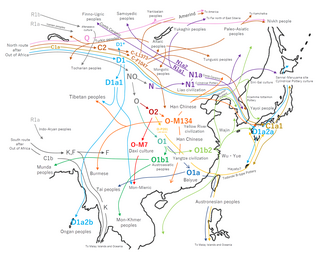
The tables below provide statistics on the human Y-chromosome DNA haplogroups most commonly found among ethnolinguistic groups and populations from East and South-East Asia.
Haplogroup C1 also known as C-F3393, is a major Y-chromosome haplogroup. It is one of two primary branches of the broader Haplogroup C, the other being C2.

Haplogroup O2a2b1a1-M117 is a subclade of O2a2b1-M134 that occurs frequently in China and in neighboring countries like Nepal, Bhutan, and Korea, especially among Sino-Tibetan language speaking people.
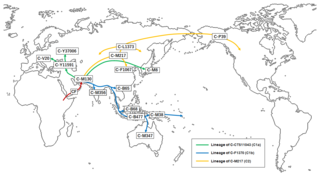
Haplogroup C-M8 also known as Haplogroup C1a1 is a Y-chromosome haplogroup. It is one of two branches of Haplogroup C1a, one of the descendants of Haplogroup C-M130.
Haplogroup D-M55 (M64.1/Page44.1) also known as Haplogroup D1a2a is a Y-chromosome haplogroup. It is one of two branches of Haplogroup D1a. The other is D1a1, which is found with high frequency in Tibetans and other Tibeto-Burmese populations and geographical close groups. D is also distributed with low to medium frequency in Central Asia, East Asia, and Mainland Southeast Asia.
This article explains the genetic makeup and population history of East Asian peoples and closely related populations, which are collectively referred to as "East-Eurasians" in population genomics.
{{cite journal}}: Cite journal requires |journal= (help){{cite journal}}: Cite journal requires |journal= (help); Missing or empty |title= (help)[ full citation needed ]{{cite journal}}: Cite journal requires |journal= (help); Missing or empty |title= (help)[ full citation needed ]{{cite journal}}: Cite journal requires |journal= (help); Missing or empty |title= (help)[ full citation needed ]{{cite journal}}: Cite journal requires |journal= (help); Missing or empty |title= (help)[ full citation needed ]{{cite journal}}: Cite journal requires |journal= (help); Missing or empty |title= (help)[ full citation needed ]{{cite journal}}: Cite journal requires |journal= (help); Missing or empty |title= (help)[ full citation needed ]