
Biochemistry or biological chemistry, is the study of chemical processes within and relating to living organisms. A sub-discipline of both chemistry and biology, biochemistry may be divided into three fields: structural biology, enzymology and metabolism. Over the last decades of the 20th century, biochemistry has become successful at explaining living processes through these three disciplines. Almost all areas of the life sciences are being uncovered and developed through biochemical methodology and research. Biochemistry focuses on understanding the chemical basis which allows biological molecules to give rise to the processes that occur within living cells and between cells, in turn relating greatly to the understanding of tissues and organs, as well as organism structure and function. Biochemistry is closely related to molecular biology, which is the study of the molecular mechanisms of biological phenomena.
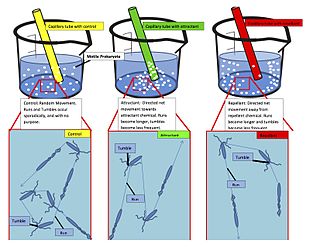
Chemotaxis is the movement of an organism or entity in response to a chemical stimulus. Somatic cells, bacteria, and other single-cell or multicellular organisms direct their movements according to certain chemicals in their environment. This is important for bacteria to find food by swimming toward the highest concentration of food molecules, or to flee from poisons. In multicellular organisms, chemotaxis is critical to early development and subsequent phases of development as well as in normal function and health. In addition, it has been recognized that mechanisms that allow chemotaxis in animals can be subverted during cancer metastasis. The aberrant chemotaxis of leukocytes and lymphocytes also contribute to inflammatory diseases such as atherosclerosis, asthma, and arthritis. Sub-cellular components, such as the polarity patch generated by mating yeast, may also display chemotactic behavior.

A hormone is any member of a class of signaling molecules in multicellular organisms, that are transported by intricate biological processes to distant organs to regulate physiology and behavior. Hormones are required for the correct development of animals, plants and fungi. The lax definition of a hormone means that many different classes of molecule can be defined as hormones. Among the substances that can be considered hormones, are eicosanoids, steroids, amino acid derivatives, protein / peptides and gases.

Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity.

A macromolecule is a very large molecule important to biophysical processes, such as a protein or nucleic acid. They are composed of thousands of covalently bonded atoms. Many macromolecules are polymers of smaller molecules called monomers. The most common macromolecules in biochemistry are biopolymers and large non-polymeric molecules such as lipids, nanogels and macrocycles. Synthetic fibers and experimental materials such as carbon nanotubes are also examples of macromolecules.

Biochemistry is the study of the chemical processes in living organisms. It deals with the structure and function of cellular components such as proteins, carbohydrates, lipids, nucleic acids and other biomolecules.

Structural bioinformatics is the branch of bioinformatics that is related to the analysis and prediction of the three-dimensional structure of biological macromolecules such as proteins, RNA, and DNA. It deals with generalizations about macromolecular 3D structures such as comparisons of overall folds and local motifs, principles of molecular folding, evolution, binding interactions, and structure/function relationships, working both from experimentally solved structures and from computational models. The term structural has the same meaning as in structural biology, and structural bioinformatics can be seen as a part of computational structural biology. The main objective of structural bioinformatics is the creation of new methods of analysing and manipulating biological macromolecular data in order to solve problems in biology and generate new knowledge.

Auxins are a class of plant hormones with some morphogen-like characteristics. Auxins play a cardinal role in coordination of many growth and behavioral processes in plant life cycles and are essential for plant body development. The Dutch biologist Frits Warmolt Went first described auxins and their role in plant growth in the 1920s. Kenneth V. Thimann became the first to isolate one of these phytohormones and to determine its chemical structure as indole-3-acetic acid (IAA). Went and Thimann co-authored a book on plant hormones, Phytohormones, in 1937.

Drug design, often referred to as rational drug design or simply rational design, is the inventive process of finding new medications based on the knowledge of a biological target. The drug is most commonly an organic small molecule that activates or inhibits the function of a biomolecule such as a protein, which in turn results in a therapeutic benefit to the patient. In the most basic sense, drug design involves the design of molecules that are complementary in shape and charge to the biomolecular target with which they interact and therefore will bind to it. Drug design frequently but not necessarily relies on computer modeling techniques. This type of modeling is sometimes referred to as computer-aided drug design. Finally, drug design that relies on the knowledge of the three-dimensional structure of the biomolecular target is known as structure-based drug design. In addition to small molecules, biopharmaceuticals including peptides and especially therapeutic antibodies are an increasingly important class of drugs and computational methods for improving the affinity, selectivity, and stability of these protein-based therapeutics have also been developed.
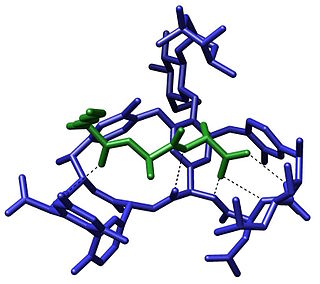
The term molecular recognition refers to the specific interaction between two or more molecules through noncovalent bonding such as hydrogen bonding, metal coordination, hydrophobic forces, van der Waals forces, π-π interactions, halogen bonding, or resonant interaction effects. In addition to these direct interactions, solvents can play a dominant indirect role in driving molecular recognition in solution. The host and guest involved in molecular recognition exhibit molecular complementarity. Exceptions are molecular containers, including e.g. nanotubes, in which portals essentially control selectivity.
Chemical biology is a scientific discipline spanning the fields of chemistry and biology. The discipline involves the application of chemical techniques, analysis, and often small molecules produced through synthetic chemistry, to the study and manipulation of biological systems. In contrast to biochemistry, which involves the study of the chemistry of biomolecules and regulation of biochemical pathways within and between cells, chemical biology deals with chemistry applied to biology.

In the field of molecular modeling, docking is a method which predicts the preferred orientation of one molecule to a second when a ligand and a target are bound to each other to form a stable complex. Knowledge of the preferred orientation in turn may be used to predict the strength of association or binding affinity between two molecules using, for example, scoring functions.

The following outline is provided as an overview of and topical guide to biochemistry:

Biomolecular structure is the intricate folded, three-dimensional shape that is formed by a molecule of protein, DNA, or RNA, and that is important to its function. The structure of these molecules may be considered at any of several length scales ranging from the level of individual atoms to the relationships among entire protein subunits. This useful distinction among scales is often expressed as a decomposition of molecular structure into four levels: primary, secondary, tertiary, and quaternary. The scaffold for this multiscale organization of the molecule arises at the secondary level, where the fundamental structural elements are the molecule's various hydrogen bonds. This leads to several recognizable domains of protein structure and nucleic acid structure, including such secondary-structure features as alpha helixes and beta sheets for proteins, and hairpin loops, bulges, and internal loops for nucleic acids. The terms primary, secondary, tertiary, and quaternary structure were introduced by Kaj Ulrik Linderstrøm-Lang in his 1951 Lane Medical Lectures at Stanford University.

Molecular biophysics is a rapidly evolving interdisciplinary area of research that combines concepts in physics, chemistry, engineering, mathematics and biology. It seeks to understand biomolecular systems and explain biological function in terms of molecular structure, structural organization, and dynamic behaviour at various levels of complexity. This discipline covers topics such as the measurement of molecular forces, molecular associations, allosteric interactions, Brownian motion, and cable theory. Additional areas of study can be found on Outline of Biophysics. The discipline has required development of specialized equipment and procedures capable of imaging and manipulating minute living structures, as well as novel experimental approaches.
Biomolecular engineering is the application of engineering principles and practices to the purposeful manipulation of molecules of biological origin. Biomolecular engineers integrate knowledge of biological processes with the core knowledge of chemical engineering in order to focus on molecular level solutions to issues and problems in the life sciences related to the environment, agriculture, energy, industry, food production, biotechnology and medicine.
Molecular binding is an attractive interaction between two molecules that results in a stable association in which the molecules are in close proximity to each other. It is formed when atoms or molecules bind together by sharing of electrons. It often, but not always, involves some chemical bonding.

Peter B. Dervan is the Bren Professor of Chemistry at the California Institute of Technology. The primary focus of his research is the development and study of small organic molecules that can sequence-specifically recognize DNA, a field in which he is an internationally recognized authority. The most important of these small molecules are pyrrole–imidazole polyamides. Dervan is credited with influencing "the course of research in organic chemistry through his studies at the interface of chemistry and biology" as a result of his work on "the chemical principles involved in sequence-specific recognition of double helical DNA". He is the recipient of many awards, including the National Medal of Science (2006).
Affibody molecules are small, robust proteins engineered to bind to a large number of target proteins or peptides with high affinity, imitating monoclonal antibodies, and are therefore a member of the family of antibody mimetics. Affibody molecules are used in biochemical research and are being developed as potential new biopharmaceutical drugs. These molecules can be used for molecular recognition in diagnostic and therapeutic applications.

Strigolactones are a group of chemical compounds produced by a plant's roots. Due to their mechanism of action, these molecules have been classified as plant hormones or phytohormones. So far, strigolactones have been identified to be responsible for three different physiological processes: First, they promote the germination of parasitic organisms that grow in the host plant's roots, such as Strigalutea and other plants of the genus Striga. Second, strigolactones are fundamental for the recognition of the plant by symbiotic fungi, especially arbuscular mycorrhizal fungi, because they establish a mutualistic association with these plants, and provide phosphate and other soil nutrients. Third, strigolactones have been identified as branching inhibition hormones in plants; when present, these compounds prevent excess bud growing in stem terminals, stopping the branching mechanism in plants.












