| Ion channel (eukaryotic) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
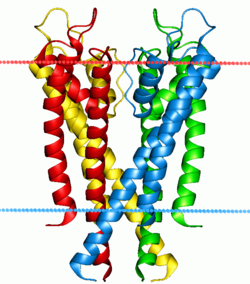
 Potassium channel Kv1.2 (with beta2 auxiliary subunits), structure in a membrane-like environment. Calculated hydrocarbon boundaries of the lipid bilayer are indicated by red and blue dots. | |||||||||||
| Identifiers | |||||||||||
| Symbol | Ion_trans | ||||||||||
| Pfam | PF00520 | ||||||||||
| InterPro | IPR005821 | ||||||||||
| SCOP2 | 1bl8 / SCOPe / SUPFAM | ||||||||||
| TCDB | 1.A.1 | ||||||||||
| OPM superfamily | 8 | ||||||||||
| OPM protein | 2a79 | ||||||||||
| |||||||||||
The transmembrane cation channel superfamily was defined in InterPro and Pfam as the family of tetrameric ion channels. These include the sodium, potassium, [1] calcium, ryanodine receptor, HCN, CNG, CatSper, and TRP channels. This large group of ion channels apparently includes families 1.A.1 , 1.A.2 , 1.A.3 , and 1.A.4 of the TCDB transporter classification.
They are described as minimally having two transmembrane helices flanking a loop which determines the ion selectivity of the channel pore. Many eukaryotic channels have four additional transmembrane helices (TM) (Pfam PF00520), related to or vestigial of voltage gating. The proteins with only two transmembrane helices (Pfam PF07885) are most commonly found in bacteria. This also includes the 2-TM inward-rectifier potassium channels (Pfam PF01007) found primarily in eukaryotes. There are commonly additional regulatory domains which serve to regulate ion conduction and channel gating. The pores may also be homotetramers or heterotetramers; where heterotetramers may be encoded as distinct genes or as multiple pore domains within a single polypeptide. The HVCN1 and Putative tyrosine-protein phosphatase proteins do not contain an expected ion conduction pore domain, but rather have homology only to the voltage sensor domain of voltage gated ion channels.