
In molecular genetics, the three prime untranslated region (3′-UTR) is the section of messenger RNA (mRNA) that immediately follows the translation termination codon. The 3′-UTR often contains regulatory regions that post-transcriptionally influence gene expression.

Picornaviruses are a group of related nonenveloped RNA viruses which infect vertebrates including fish, mammals, and birds. They are viruses that represent a large family of small, positive-sense, single-stranded RNA viruses with a 30 nm icosahedral capsid. The viruses in this family can cause a range of diseases including the common cold, poliomyelitis, meningitis, hepatitis, and paralysis.
The 5′ untranslated region is the region of a messenger RNA (mRNA) that is directly upstream from the initiation codon. This region is important for the regulation of translation of a transcript by differing mechanisms in viruses, prokaryotes and eukaryotes. While called untranslated, the 5′ UTR or a portion of it is sometimes translated into a protein product. This product can then regulate the translation of the main coding sequence of the mRNA. In many organisms, however, the 5′ UTR is completely untranslated, instead forming a complex secondary structure to regulate translation.
VPg is a protein that is covalently attached to the 5′ end of positive strand viral RNA and acts as a primer during RNA synthesis in a variety of virus families including Picornaviridae, Potyviridae, Astroviridae and Caliciviridae. There are some studies showing that a possible VPg protein is also present in astroviridae, however, experimental evidence for this has not yet been provided and requires further study. The primer activity of VPg occurs when the protein becomes uridylated, providing a free hydroxyl that can be extended by the virally encoded RNA-dependent RNA polymerase. For some virus families, VPg also has a role in translation initiation by acting like a 5' mRNA cap.
Ribosome shunting is a mechanism of translation initiation in which ribosomes bypass, or "shunt over", parts of the 5' untranslated region to reach the start codon. However, a benefit of ribosomal shunting is that it can translate backwards allowing more information to be stored than usual in an mRNA molecule. Some viral RNAs have been shown to use ribosome shunting as a more efficient form of translation during certain stages of viral life cycle or when translation initiation factors are scarce. Some viruses known to use this mechanism include adenovirus, Sendai virus, human papillomavirus, duck hepatitis B pararetrovirus, rice tungro bacilliform viruses, and cauliflower mosaic virus. In these viruses the ribosome is directly translocated from the upstream initiation complex to the start codon (AUG) without the need to unwind RNA secondary structures.

In molecular biology, the coronavirus frameshifting stimulation element is a conserved stem-loop of RNA found in coronaviruses that can promote ribosomal frameshifting. Such RNA molecules interact with a downstream region to form a pseudoknot structure; the region varies according to the virus but pseudoknot formation is known to stimulate frameshifting. In the classical situation, a sequence 32 nucleotides downstream of the stem is complementary to part of the loop. In other coronaviruses, however, another stem-loop structure around 150 nucleotides downstream can interact with members of this family to form kissing stem-loops and stimulate frameshifting.

The Coronavirus packaging signal is a conserved cis-regulatory element found in Betacoronavirus. It has an important role in regulating the packaging of the viral genome into the capsid. As part of the viral life cycle, within the infected cell, the viral genome becomes associated with viral proteins and assembles into new infective progeny viruses. This process is called packaging and is vital for viral replication.

This family represents the internal ribosome entry site (IRES) of the hepatitis A virus. HAV IRES is a 450 nucleotide long sequence located in the 735 nt long 5’ UTR of Hepatitis A viral RNA genome. IRES elements allow cap and end-independent translation of mRNA in the host cell. The IRES achieves this by mediating the internal initiation of translation by recruiting a ribosomal 40S pre-initiation complex directly to the initiation codon and eliminates the requirement for eukaryotic initiation factor, eIF4F.
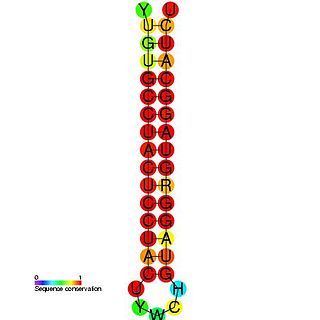
The Hepatitis C stem-loop IV is part of a putative RNA element found in the NS5B coding region. This element along with stem-loop VII, is important for colony formation, though its exact function and mechanism are unknown.

The hepatitis C virus 3′X element is an RNA element which contains three stem-loop structures that are essential for replication.

The Hepatitis C virus (HCV) cis-acting replication element (CRE) is an RNA element which is found in the coding region of the RNA-dependent RNA polymerase NS5B. Mutations in this family have been found to cause a blockage in RNA replication and it is thought that both the primary sequence and the structure of this element are crucial for HCV RNA replication.
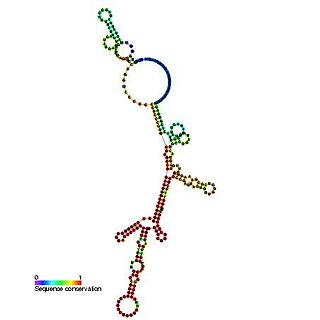
The Hepatitis C virus internal ribosome entry site, or HCV IRES, is an RNA structure within the 5'UTR of the HCV genome that mediates cap-independent translation initiation.
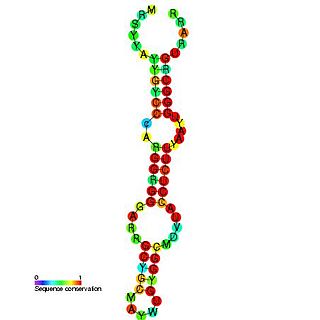
Hepatitis C virus stem-loop VII is a regulatory element found in the coding region of the RNA-dependent RNA polymerase gene, NS5B. Similarly to stem-loop IV, the stem-loop structure is important for colony formation, though its exact function and mechanism are unknown.
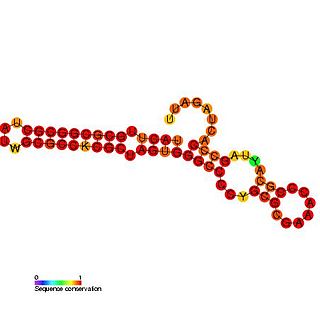
The Rubella virus 3′ cis-acting element RNA family represents a cis-acting element found at the 3′ UTR in the rubella virus. This family contains three conserved step loop structures. Calreticulin (CAL), which is known to bind calcium in most eukaryotic cells, is able to specifically bind to the first stem loop of this RNA. CAL binding is thought to be related to viral pathogenesis and in particular arthritis which occurs frequently in rubella infections in adults and is independent of viral viability. All stem loop structures are thought to be important for efficient viral replication and deletion of stem loop three is known to be lethal.
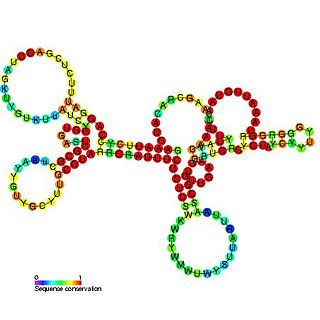
Tombusvirus 5′ UTR is an important cis-regulatory region of the Tombus virus genome.

Hepatitis C alternative reading frame stem-loop is a conserved secondary structure motif identified in the RNA genome of the hepatitis C virus (HCV) which is proposed to have an important role in regulating translation and repression of the viral genome.
Ribosomal frameshifting, also known as translational frameshifting or translational recoding, is a biological phenomenon that occurs during translation that results in the production of multiple, unique proteins from a single mRNA. The process can be programmed by the nucleotide sequence of the mRNA and is sometimes affected by the secondary, 3-dimensional mRNA structure. It has been described mainly in viruses, retrotransposons and bacterial insertion elements, and also in some cellular genes.

Red clover necrotic mosaic virus (RCNMV) contains several structural elements present within the 3′ and 5′ untranslated regions (UTR) of the genome that enhance translation. In eukaryotes transcription is a prerequisite for translation. During transcription the pre-mRNA transcript is processes where a 5′ cap is attached onto mRNA and this 5′ cap allows for ribosome assembly onto the mRNA as it acts as a binding site for the eukaryotic initiation factor eIF4F. Once eIF4F is bound to the mRNA this protein complex interacts with the poly(A) binding protein which is present within the 3′ UTR and results in mRNA circularization. This multiprotein-mRNA complex then recruits the ribosome subunits and scans the mRNA until it reaches the start codon. Transcription of viral genomes differs from eukaryotes as viral genomes produce mRNA transcripts that lack a 5’ cap site. Despite lacking a cap site viral genes contain a structural element within the 5’ UTR known as an internal ribosome entry site (IRES). IRES is a structural element that recruits the 40s ribosome subunit to the mRNA within close proximity of the start codon.

The rmf RNA motif is a conserved RNA structure that was originally detected using bioinformatics. rmf RNAs are consistently foundwithin species classified into the genus Pseudomonas, and is located potentially in the 5′ untranslated regions of rmf genes. These genes encodes the ribosome modulation factor protein, which affects the translation of genes by modifying ribosome structure in response to stress such as starvation. This ribosome modulation is a part of the stringent response in bacteria. The likely biological role of rmf RNAs is ambiguous. Since the RNA could be in the 5′ UTRs of protein-coding genes, it was hypothesized that it functions as a cis-regulatory element. This hypothesis is bolstered by the observation that ribosome modulation factor binds ribosomal RNA, and many cis-regulatory RNAs called ribosomal protein leaders participate in a feedback regulation mechanism by binding to proteins that normally bind to ribosomal RNA. However, since rmf RNAs are not very close to the rmf genes, they might function as non-coding RNAs.
Cis-acting replication elements bring together the 5′ and 3′ ends during replication of positive-sense single-stranded RNA viruses and double-stranded RNA viruses.