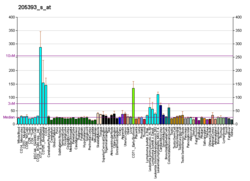
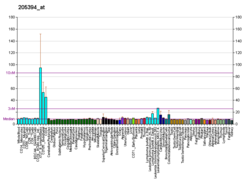
Checkpoint kinase 1, commonly referred to as Chk1, is a serine/threonine-specific protein kinase that, in humans, is encoded by the CHEK1 gene. [5] [6] Chk1 coordinates the DNA damage response (DDR) and cell cycle checkpoint response. [7] Activation of Chk1 results in the initiation of cell cycle checkpoints, cell cycle arrest, DNA repair and cell death to prevent damaged cells from progressing through the cell cycle.