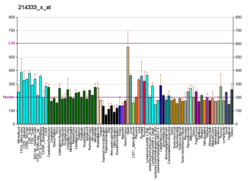
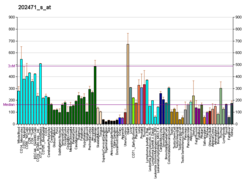
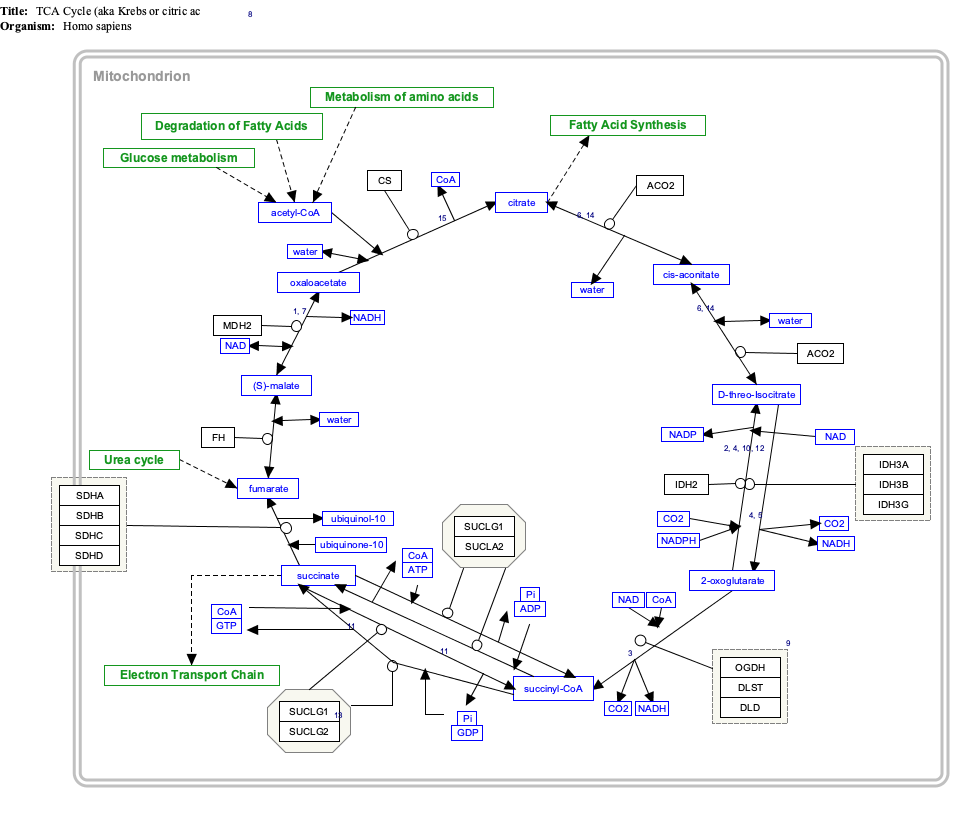
Isocitrate dehydrogenase (IDH) (EC 1.1.1.42) and (EC 1.1.1.41) is an enzyme that catalyzes the oxidative decarboxylation of isocitrate, producing alpha-ketoglutarate (α-ketoglutarate) and CO2. This is a two-step process, which involves oxidation of isocitrate (a secondary alcohol) to oxalosuccinate (a ketone), followed by the decarboxylation of the carboxyl group beta to the ketone, forming alpha-ketoglutarate. In humans, IDH exists in three isoforms: IDH3 catalyzes the third step of the citric acid cycle while converting NAD+ to NADH in the mitochondria. The isoforms IDH1 and IDH2 catalyze the same reaction outside the context of the citric acid cycle and use NADP+ as a cofactor instead of NAD+. They localize to the cytosol as well as the mitochondrion and peroxisome.
Isocitrate dehydrogenase (NAD+) (EC 1.1.1.41, isocitric dehydrogenase, beta-ketoglutaric-isocitric carboxylase, isocitric acid dehydrogenase, NAD dependent isocitrate dehydrogenase, NAD isocitrate dehydrogenase, NAD-linked isocitrate dehydrogenase, NAD-specific isocitrate dehydrogenase, NAD isocitric dehydrogenase, isocitrate dehydrogenase (NAD), IDH (ambiguous), nicotinamide adenine dinucleotide isocitrate dehydrogenase) is an enzyme with systematic name isocitrate:NAD+ oxidoreductase (decarboxylating). This enzyme catalyses the following chemical reaction

Trifunctional enzyme subunit alpha, mitochondrial also known as hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase, alpha subunit is a protein that in humans is encoded by the HADHA gene. Mutations in HADHA have been associated with trifunctional protein deficiency or long-chain 3-hydroxyacyl-coenzyme A dehydrogenase deficiency.

Trifunctional enzyme subunit beta, mitochondrial (TP-beta) also known as 3-ketoacyl-CoA thiolase, acetyl-CoA acyltransferase, or beta-ketothiolase is an enzyme that in humans is encoded by the HADHB gene.

Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 is a protein that in humans is encoded by the GNB1 gene.

5'-AMP-activated protein kinase subunit gamma-2 is an enzyme that in humans is encoded by the PRKAG2 gene.

Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-2 is a protein that in humans is encoded by the GNB2 gene.

Eukaryotic translation initiation factor 2 subunit 2 (eIF2β) is a protein that in humans is encoded by the EIF2S2 gene.

Phosphoglycerate dehydrogenase (PHGDH) is an enzyme that catalyzes the chemical reactions

2-Oxoisovalerate dehydrogenase subunit beta, mitochondrial is an enzyme that in humans is encoded by the BCKDHB gene.

Voltage-gated potassium channel subunit beta-1 is a protein that in humans is encoded by the KCNAB1 gene.

Guanine nucleotide-binding protein G(T) subunit gamma-T1 is a protein that in humans is encoded by the GNGT1 gene.

Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial (IDH3α) is an enzyme that in humans is encoded by the IDH3A gene.

Isocitrate dehydrogenase [NADP], mitochondrial is an enzyme that in humans is encoded by the IDH2 gene.

Voltage-dependent L-type calcium channel subunit beta-3 is a protein that in humans is encoded by the CACNB3 gene.

Isocitrate dehydrogenase [NAD] subunit beta, mitochondrial is an enzyme that in humans is encoded by the IDH3B gene.

Malate dehydrogenase, mitochondrial also known as malate dehydrogenase 2 is an enzyme that in humans is encoded by the MDH2 gene.

Pyruvate dehydrogenase (lipoamide) alpha 2, also known as pyruvate dehydrogenase E1 component subunit alpha, testis-specific form, mitochondrial or PDHE1-A type II, is an enzyme that in humans is encoded by the PDHA2 gene.

Pyruvate dehydrogenase (lipoamide) beta, also known as pyruvate dehydrogenase E1 component subunit beta, mitochondrial or PDHE1-B is an enzyme that in humans is encoded by the PDHB gene. The pyruvate dehydrogenase (PDH) complex is a nuclear-encoded mitochondrial multienzyme complex that catalyzes the overall conversion of pyruvate to acetyl-CoA and CO2, and provides the primary link between glycolysis and the tricarboxylic acid (TCA) cycle. The PDH complex is composed of multiple copies of three enzymatic components: pyruvate dehydrogenase (E1), dihydrolipoamide acetyltransferase (E2) and lipoamide dehydrogenase (E3). The E1 enzyme is a heterotetramer of two alpha and two beta subunits. This gene encodes the E1 beta subunit. Mutations in this gene are associated with pyruvate dehydrogenase E1-beta deficiency.

Isocitrate dehydrogenase 1 (NADP+), soluble is an enzyme that in humans is encoded by the IDH1 gene on chromosome 2. Isocitrate dehydrogenases catalyze the oxidative decarboxylation of isocitrate to 2-oxoglutarate. These enzymes belong to two distinct subclasses, one of which uses NAD+ as the electron acceptor and the other NADP+. Five isocitrate dehydrogenases have been reported: three NAD+-dependent isocitrate dehydrogenases, which localize to the mitochondrial matrix, and two NADP+-dependent isocitrate dehydrogenases, one of which is mitochondrial and the other predominantly cytosolic. Each NADP+-dependent isozyme is a homodimer. The protein encoded by this gene is the NADP+-dependent isocitrate dehydrogenase found in the cytoplasm and peroxisomes. It contains the PTS-1 peroxisomal targeting signal sequence. The presence of this enzyme in peroxisomes suggests roles in the regeneration of NADPH for intraperoxisomal reductions, such as the conversion of 2,4-dienoyl-CoAs to 3-enoyl-CoAs, as well as in peroxisomal reactions that consume 2-oxoglutarate, namely the alpha-hydroxylation of phytanic acid. The cytoplasmic enzyme serves a significant role in cytoplasmic NADPH production. Alternatively spliced transcript variants encoding the same protein have been found for this gene. [provided by RefSeq, Sep 2013]