
The citric acid cycle —also known as the Krebs cycle, Szent-Györgyi-Krebs cycle or the TCA cycle (tricarboxylic acid cycle)—is a series of chemical reactions to release stored energy through the oxidation of acetyl-CoA derived from carbohydrates, fats, and proteins. The Krebs cycle is used by organisms that respire (as opposed to organisms that ferment) to generate energy, either by anaerobic respiration or aerobic respiration. In addition, the cycle provides precursors of certain amino acids, as well as the reducing agent NADH, that are used in numerous other reactions. Its central importance to many biochemical pathways suggests that it was one of the earliest components of metabolism. Even though it is branded as a 'cycle', it is not necessary for metabolites to follow only one specific route; at least three alternative segments of the citric acid cycle have been recognized.
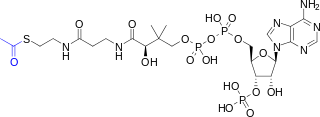
Acetyl-CoA is a molecule that participates in many biochemical reactions in protein, carbohydrate and lipid metabolism. Its main function is to deliver the acetyl group to the citric acid cycle to be oxidized for energy production. Coenzyme A consists of a β-mercaptoethylamine group linked to the vitamin pantothenic acid (B5) through an amide linkage and 3'-phosphorylated ADP. The acetyl group of acetyl-CoA is linked to the sulfhydryl substituent of the β-mercaptoethylamine group. This thioester linkage is a "high energy" bond, which is particularly reactive. Hydrolysis of the thioester bond is exergonic (−31.5 kJ/mol).

Pyruvate kinase is the enzyme involved in the last step of glycolysis. It catalyzes the transfer of a phosphate group from phosphoenolpyruvate (PEP) to adenosine diphosphate (ADP), yielding one molecule of pyruvate and one molecule of ATP. Pyruvate kinase was inappropriately named before it was recognized that it did not directly catalyze phosphorylation of pyruvate, which does not occur under physiological conditions. Pyruvate kinase is present in four distinct, tissue-specific isozymes in animals, each consisting of particular kinetic properties necessary to accommodate the variations in metabolic requirements of diverse tissues.

Pyruvate dehydrogenase complex (PDC) is a complex of three enzymes that converts pyruvate into acetyl-CoA by a process called pyruvate decarboxylation. Acetyl-CoA may then be used in the citric acid cycle to carry out cellular respiration, and this complex links the glycolysis metabolic pathway to the citric acid cycle. Pyruvate decarboxylation is also known as the "pyruvate dehydrogenase reaction" because it also involves the oxidation of pyruvate.

In the mitochondrion, the matrix is the space within the inner membrane. The word "matrix" stems from the fact that this space is viscous, compared to the relatively aqueous cytoplasm. The mitochondrial matrix contains the mitochondrial DNA, ribosomes, soluble enzymes, small organic molecules, nucleotide cofactors, and inorganic ions.[1] The enzymes in the matrix facilitate reactions responsible for the production of ATP, such as the citric acid cycle, oxidative phosphorylation, oxidation of pyruvate, and the beta oxidation of fatty acids.
Pyruvate dehydrogenase lipoamide kinase isozyme 1, mitochondrial is an enzyme that in humans is encoded by the PDK1 gene. It codes for an isozyme of pyruvate dehydrogenase kinase (PDK).
Phosphoenolpyruvate carboxylase (also known as PEP carboxylase, PEPCase, or PEPC; EC 4.1.1.31, PDB ID: 3ZGE) is an enzyme in the family of carboxy-lyases found in plants and some bacteria that catalyzes the addition of bicarbonate (HCO3−) to phosphoenolpyruvate (PEP) to form the four-carbon compound oxaloacetate and inorganic phosphate:
Pyruvate dehydrogenase is an enzyme that catalyzes the reaction of pyruvate and a lipoamide to give the acetylated dihydrolipoamide and carbon dioxide. The conversion requires the coenzyme thiamine pyrophosphate.

Dihydrolipoamide dehydrogenase (DLD), also known as dihydrolipoyl dehydrogenase, mitochondrial, is an enzyme that in humans is encoded by the DLD gene. DLD is a flavoprotein enzyme that oxidizes dihydrolipoamide to lipoamide.

E3 binding protein also known as pyruvate dehydrogenase protein X component, mitochondrial is a protein that in humans is encoded by the PDHX gene. The E3 binding protein is a component of the pyruvate dehydrogenase complex found only in eukaryotes. Defects in this gene are a cause of pyruvate dehydrogenase deficiency which results in neurological dysfunction and lactic acidosis in infancy and early childhood. This protein is also a minor antigen for antimitochondrial antibodies. These autoantibodies are present in nearly 95% of patients with primary biliary cholangitis, an autoimmune disease of the liver. In primary biliary cholangitis, activated T lymphocytes attack and destroy epithelial cells in the bile duct where this protein is abnormally distributed and overexpressed. Primary biliary cholangitis eventually leads to liver failure.
The PHLPP isoforms are a pair of protein phosphatases, PHLPP1 and PHLPP2, that are important regulators of Akt serine-threonine kinases and conventional/novel protein kinase C (PKC) isoforms. PHLPP may act as a tumor suppressor in several types of cancer due to its ability to block growth factor-induced signaling in cancer cells.

Pyruvate dehydrogenase phosphatase catalytic subunit 1, also known as protein phosphatase 2C, is an enzyme that in humans is encoded by the PDP1 gene. PDPC 1 is an enzyme which serves to reverse the effects of pyruvate dehydrogenase kinase upon pyruvate dehydrogenase, activating pyruvate dehydrogenase.

Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial is an enzyme that in humans is encoded by the PDHA1 gene.The pyruvate dehydrogenase complex is a nuclear-encoded mitochondrial matrix multienzyme complex that provides the primary link between glycolysis and the tricarboxylic acid (TCA) cycle by catalyzing the irreversible conversion of pyruvate into acetyl-CoA. The PDH complex is composed of multiple copies of 3 enzymes: E1 (PDHA1); dihydrolipoyl transacetylase (DLAT) ; and dihydrolipoyl dehydrogenase (DLD). The E1 enzyme is a heterotetramer of 2 alpha and 2 beta subunits. The E1-alpha subunit contains the E1 active site and plays a key role in the function of the PDH complex.
Pyruvate dehydrogenase lipoamide kinase isozyme 4, mitochondrial (PDK4) is an enzyme that in humans is encoded by the PDK4 gene. It codes for an isozyme of pyruvate dehydrogenase kinase.

Pyruvate dehydrogenase kinase isoform 2 (PDK2) also known as pyruvate dehydrogenase lipoamide kinase isozyme 2, mitochondrial is an enzyme that in humans is encoded by the PDK2 gene. PDK2 is an isozyme of pyruvate dehydrogenase kinase.

Pyruvate dehydrogenase lipoamide kinase isozyme 3, mitochondrial is an enzyme that in humans is encoded by the PDK3 gene. It codes for an isozyme of pyruvate dehydrogenase kinase.The pyruvate dehydrogenase (PDH) complex is a nuclear-encoded mitochondrial multienzyme complex that catalyzes the overall conversion of pyruvate to acetyl-CoA and CO2. It provides the primary link between glycolysis and the tricarboxylic acid (TCA) cycle, and thus is one of the major enzymes responsible for the regulation of glucose metabolism. The enzymatic activity of PDH is regulated by a phosphorylation/dephosphorylation cycle, and phosphorylation results in inactivation of PDH. The protein encoded by this gene is one of the four pyruvate dehydrogenase kinases that inhibits the PDH complex by phosphorylation of the E1 alpha subunit. This gene is predominantly expressed in the heart and skeletal muscles. Alternatively spliced transcript variants encoding different isoforms have been found for this gene.
Glutaminolysis (glutamine + -lysis) is a series of biochemical reactions by which the amino acid glutamine is lysed to glutamate, aspartate, CO2, pyruvate, lactate, alanine and citrate.

Pyruvate kinase isozymes M1/M2 (PKM1/M2), also known as pyruvate kinase muscle isozyme (PKM), pyruvate kinase type K, cytosolic thyroid hormone-binding protein (CTHBP), thyroid hormone-binding protein 1 (THBP1), or opa-interacting protein 3 (OIP3), is an enzyme that in humans is encoded by the PKM2 gene.

In the field of biochemistry, PDPK1 refers to the protein 3-phosphoinositide-dependent protein kinase-1, an enzyme which is encoded by the PDPK1 gene in humans. It is implicated in the development and progression of melanomas.

Pyruvate dehydrogenase (lipoamide) alpha 2, also known as pyruvate dehydrogenase E1 component subunit alpha, testis-specific form, mitochondrial or PDHE1-A type II, is an enzyme that in humans is encoded by the PDHA2 gene.


















