
In a neuron, synaptic vesicles store various neurotransmitters that are released at the synapse. The release is regulated by a voltage-dependent calcium channel. Vesicles are essential for propagating nerve impulses between neurons and are constantly recreated by the cell. The area in the axon that holds groups of vesicles is an axon terminal or "terminal bouton". Up to 130 vesicles can be released per bouton over a ten-minute period of stimulation at 0.2 Hz. In the visual cortex of the human brain, synaptic vesicles have an average diameter of 39.5 nanometers (nm) with a standard deviation of 5.1 nm.

SNARE proteins – "SNAPREceptors" – are a large protein family consisting of at least 24 members in yeasts, more than 60 members in mammalian cells, and some numbers in plants. The primary role of SNARE proteins is to mediate the fusion of vesicles with the target membrane; this notably mediates exocytosis, but can also mediate the fusion of vesicles with membrane-bound compartments. The best studied SNAREs are those that mediate the release of synaptic vesicles containing neurotransmitters in neurons. These neuronal SNAREs are the targets of the neurotoxins responsible for botulism and tetanus produced by certain bacteria.
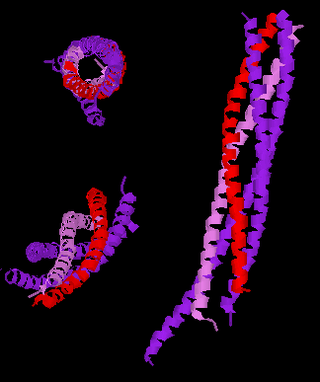
Synaptobrevins are small integral membrane proteins of secretory vesicles with molecular weight of 18 kilodalton (kDa) that are part of the vesicle-associated membrane protein (VAMP) family.

Syntaxin-1A is a protein that in humans is encoded by the STX1A gene.

Synaptosomal-associated protein 23 is a protein that in humans is encoded by the SNAP23 gene. Two alternative transcript variants encoding different protein isoforms have been described for this gene.

Syntaxin-4 is a protein that in humans is encoded by the STX4 gene.

Synaptotagmin-1 is a protein that in humans is encoded by the SYT1 gene.

Vesicle-associated membrane protein 2 (VAMP2) is a protein that in humans is encoded by the VAMP2 gene.

Syntaxin-binding protein 1 is a protein that in humans is encoded by the STXBP1 gene. This gene encodes a syntaxin-binding protein. The encoded protein appears to play a role in release of neurotransmitters via regulation of syntaxin, a transmembrane attachment protein receptor. Mutations in this gene have been associated with neurological disorders including epilepsy, intellectual disability, and movement disorders.

Syntaxin-7 is a protein that in humans is encoded by the STX7 gene.

Syntaxin-6 is a protein that in humans is encoded by the STX6 gene.

Vesicle-associated membrane protein 3 is a protein that in humans is encoded by the VAMP3 gene.

Syntaxin-5 is a protein that in humans is encoded by the STX5 gene.

Syntaxin-2, also known as epimorphin, is a protein that in humans is encoded by the STX2 gene.

Vesicle-associated membrane protein 4 is a protein that in humans is encoded by the VAMP4 gene.

Vesicle-associated membrane protein 1 (VAMP1) is a protein that in humans is encoded by the VAMP1 gene.

Syntaxin 3, also known as STX3, is a protein which in humans is encoded by the STX3 gene.
Munc-18 proteins are the mammalian homologue of UNC-18 and are a member of the Sec1/Munc18-like (SM) protein family. Munc-18 proteins have been identified as essential components of the synaptic vesicle fusion protein complex and are crucial for the regulated exocytosis of neurons and neuroendocrine cells.

Soluble N-ethylmaleimide-Sensitive Factor Attachment Proteins are a family of cytosolic adaptor proteins involved in vesicular fusion at membranes during intracellular transport and exocytosis. SNAPs interact with proteins of the SNARE complex and NSF to play a key role in recycling the components of the fusion complex. SNAPs are involved in the priming of the vesicle fusion complex during assembly, as well as in the disassembly following a vesicle fusion event. Following membrane fusion, the tethering SNARE proteins complex disassembles in response to steric changes originating from the ATPase NSF. The energy provided by NSF is transferred throughout the SNARE complex and SNAP, allowing the proteins to untangle, and recycled for future fusion events. Mammals have three SNAP genes: α-SNAP, β-SNAP, and γ-SNAP. α- and γ-SNAP are expressed throughout the body, while β-SNAP is specific to the brain. The yeast homolog of the human SNAP is Sec17, the structural diagram of which is included on this page.
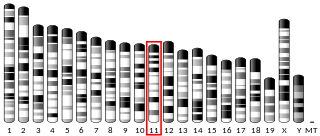
Synaptosome-associated protein, 47 kDal (SNAP47) is a human protein encoded by the SNAP47 gene. Other aliases of this gene are SVAP1, HEL170, ESFI5812, and HEL-S-290. SNAP47 is a synaptosome protein which is associated with the protein coding in multiple diseases, including non small cell lung cancer and schizophrenia. SNAP47 is a member of the SNAP protein family. SNAP proteins are t-snare proteins that are a component of SNARE complex. The SNARE complex mediates vesicle fusion by creating tight complex that brings vesicle and membrane together. This protein causes ubiquitous expression in testis, ovary, and many other tissues






























