
Glycolysis is the metabolic pathway that converts glucose into pyruvate and, in most organisms, occurs in the liquid part of cells. The free energy released in this process is used to form the high-energy molecules adenosine triphosphate (ATP) and reduced nicotinamide adenine dinucleotide (NADH). Glycolysis is a sequence of ten reactions catalyzed by enzymes.

Nucleotides are organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver.

In biochemistry, a kinase is an enzyme that catalyzes the transfer of phosphate groups from high-energy, phosphate-donating molecules to specific substrates. This process is known as phosphorylation, where the high-energy ATP molecule donates a phosphate group to the substrate molecule. As a result, kinase produces a phosphorylated substrate and ADP. Conversely, it is referred to as dephosphorylation when the phosphorylated substrate donates a phosphate group and ADP gains a phosphate group. These two processes, phosphorylation and dephosphorylation, occur four times during glycolysis.

Butyric acid, also known under the systematic name butanoic acid, is a straight-chain alkyl carboxylic acid with the chemical formula CH3CH2CH2CO2H. It is an oily, and colorless liquid with an unpleasant odor. Isobutyric acid is an isomer. Salts and esters of butyric acid are known as butyrates or butanoates. The acid does not occur widely in nature, but its esters are widespread. It is a common industrial chemical and an important component in the mammalian gut.
Gluconeogenesis (GNG) is a metabolic pathway that results in the biosynthesis of glucose from certain non-carbohydrate carbon substrates. It is a ubiquitous process, present in plants, animals, fungi, bacteria, and other microorganisms. In vertebrates, gluconeogenesis occurs mainly in the liver and, to a lesser extent, in the cortex of the kidneys. It is one of two primary mechanisms – the other being degradation of glycogen (glycogenolysis) – used by humans and many other animals to maintain blood sugar levels, avoiding low levels (hypoglycemia). In ruminants, because dietary carbohydrates tend to be metabolized by rumen organisms, gluconeogenesis occurs regardless of fasting, low-carbohydrate diets, exercise, etc. In many other animals, the process occurs during periods of fasting, starvation, low-carbohydrate diets, or intense exercise.

In biochemistry, a ribonucleotide is a nucleotide containing ribose as its pentose component. It is considered a molecular precursor of nucleic acids. Nucleotides are the basic building blocks of DNA and RNA. Ribonucleotides themselves are basic monomeric building blocks for RNA. Deoxyribonucleotides, formed by reducing ribonucleotides with the enzyme ribonucleotide reductase (RNR), are essential building blocks for DNA. There are several differences between DNA deoxyribonucleotides and RNA ribonucleotides. Successive nucleotides are linked together via phosphodiester bonds.
A nucleoside triphosphate is a nucleoside containing a nitrogenous base bound to a 5-carbon sugar, with three phosphate groups bound to the sugar. They are the molecular precursors of both DNA and RNA, which are chains of nucleotides made through the processes of DNA replication and transcription. Nucleoside triphosphates also serve as a source of energy for cellular reactions and are involved in signalling pathways.
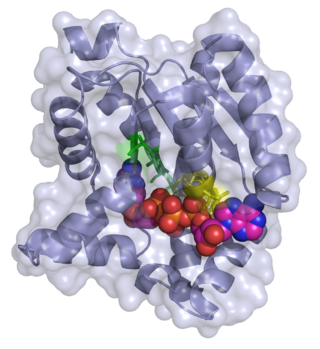
Adenylate kinase is a phosphotransferase enzyme that catalyzes the interconversion of the various adenosine phosphates. By constantly monitoring phosphate nucleotide levels inside the cell, ADK plays an important role in cellular energy homeostasis.

Nucleoside-diphosphate kinases are enzymes that catalyze the exchange of terminal phosphate between different nucleoside diphosphates (NDP) and triphosphates (NTP) in a reversible manner to produce nucleotide triphosphates. Many NDP serve as acceptor while NTP are donors of phosphate group. The general reaction via ping-pong mechanism is as follows: XDP + YTP ←→ XTP + YDP. NDPK activities maintain an equilibrium between the concentrations of different nucleoside triphosphates such as, for example, when guanosine triphosphate (GTP) produced in the citric acid (Krebs) cycle is converted to adenosine triphosphate (ATP). Other activities include cell proliferation, differentiation and development, signal transduction, G protein-coupled receptor, endocytosis, and gene expression.

Nucleic acid metabolism is a collective term that refers to the variety of chemical reactions by which nucleic acids are either synthesized or degraded. Nucleic acids are polymers made up of a variety of monomers called nucleotides. Nucleotide synthesis is an anabolic mechanism generally involving the chemical reaction of phosphate, pentose sugar, and a nitrogenous base. Degradation of nucleic acids is a catabolic reaction and the resulting parts of the nucleotides or nucleobases can be salvaged to recreate new nucleotides. Both synthesis and degradation reactions require multiple enzymes to facilitate the event. Defects or deficiencies in these enzymes can lead to a variety of diseases.

In enzymology, a nicotinate-nucleotide-dimethylbenzimidazole phosphoribosyltransferase is an enzyme that catalyzes the chemical reaction

In enzymology, an adenosylcobinamide-phosphate guanylyltransferase is an enzyme that catalyzes the chemical reaction

In enzymology, a butyrate kinase is an enzyme that catalyzes the chemical reaction
In enzymology, a glucose-1-phosphate cytidylyltransferase is an enzyme that catalyzes the chemical reaction

In enzymology, a hydroxyethylthiazole kinase is an enzyme that catalyzes the chemical reaction
In enzymology, a [isocitrate dehydrogenase (NADP+)] kinase (EC 2.7.11.5) is an enzyme that catalyzes the chemical reaction:
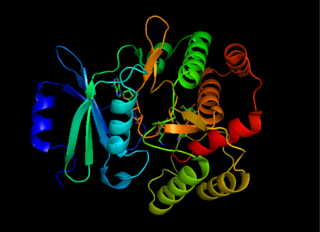
Aminoglycoside-3'-phosphotransferase, also known as aminoglycoside kinase, is an enzyme that primarily catalyzes the addition of phosphate from ATP to the 3'-hydroxyl group of a 4,6-disubstituted aminoglycoside, such as kanamycin. However, APH(3') has also been found to phosphorylate at the 5'-hydroxyl group in 4,5-disubstituted aminoglycosides, which lack a 3'-hydroxyl group, and to diphosphorylate hydroxyl groups in aminoglycosides that have both 3'- and 5'-hydroxyl groups. Primarily positively charged at biological conditions, aminoglycosides bind to the negatively charged backbone of nucleic acids to disrupt protein synthesis, effectively inhibiting bacterial cell growth. APH(3') mediated phosphorylation of aminoglycosides effectively disrupts their mechanism of action, introducing a phosphate group that reduces their binding affinity due to steric hindrances and unfavorable electrostatic interactions. APH(3') is primarily found in certain species of gram-positive bacteria.
In molecular biology, the PyrD leader is a cis-regulatory RNA element found at the 5' of the PyrC mRNA in Pseudomonadota. The PyrD gene encodes dihydroorotate dehydrogenase, an enzyme involved in pyrimidine biosynthesis. The PyrD leader regulates expression of PyrD. Translation initiation can occur at more than one different site within this leader sequence, under high cytidine triphosphate or guanosine triphosphate conditions the translation initiation site is upstream of that used under low CTP/GTP conditions, additional cytosine residues are incorporated into the mRNA resulting in the formation of an RNA hairpin. This hairpin blocks ribosome-binding at the Shine-Dalgarno sequence, and therefore blocks expression of PyrD. Under low CTP/GTP conditions the initiation site is further downstream and does not result in hairpin formation, so the ribosome can bind to the Shine-Dalgarno sequence and PyrD is expressed.
D-glycero-beta-D-manno-heptose-7-phosphate kinase is an enzyme with systematic name ATP:D-glycero-beta-D-manno-heptose 7-phosphate 1-phosphotransferase. This enzyme catalyses the following chemical reaction
Propionate kinase is an enzyme with systematic name ATP:propanoate phosphotransferase. This enzyme catalyses the following chemical reaction













