Related Research Articles

Microtubules are polymers of tubulin that form part of the cytoskeleton and provide structure and shape to eukaryotic cells. Microtubules can be as long as 50 micrometres, as wide as 23 to 27 nm and have an inner diameter between 11 and 15 nm. They are formed by the polymerization of a dimer of two globular proteins, alpha and beta tubulin into protofilaments that can then associate laterally to form a hollow tube, the microtubule. The most common form of a microtubule consists of 13 protofilaments in the tubular arrangement.

The cytoskeleton is a complex, dynamic network of interlinking protein filaments present in the cytoplasm of all cells, including those of bacteria and archaea. In eukaryotes, it extends from the cell nucleus to the cell membrane and is composed of similar proteins in the various organisms. It is composed of three main components: microfilaments, intermediate filaments, and microtubules, and these are all capable of rapid growth or disassembly depending on the cell's requirements.

Microfilaments, also called actin filaments, are protein filaments in the cytoplasm of eukaryotic cells that form part of the cytoskeleton. They are primarily composed of polymers of actin, but are modified by and interact with numerous other proteins in the cell. Microfilaments are usually about 7 nm in diameter and made up of two strands of actin. Microfilament functions include cytokinesis, amoeboid movement, cell motility, changes in cell shape, endocytosis and exocytosis, cell contractility, and mechanical stability. Microfilaments are flexible and relatively strong, resisting buckling by multi-piconewton compressive forces and filament fracture by nanonewton tensile forces. In inducing cell motility, one end of the actin filament elongates while the other end contracts, presumably by myosin II molecular motors. Additionally, they function as part of actomyosin-driven contractile molecular motors, wherein the thin filaments serve as tensile platforms for myosin's ATP-dependent pulling action in muscle contraction and pseudopod advancement. Microfilaments have a tough, flexible framework which helps the cell in movement.

Actin is a family of globular multi-functional proteins that form microfilaments in the cytoskeleton, and the thin filaments in muscle fibrils. It is found in essentially all eukaryotic cells, where it may be present at a concentration of over 100 μM; its mass is roughly 42 kDa, with a diameter of 4 to 7 nm.
MreB is a protein found in bacteria that has been identified as a homologue of actin, as indicated by similarities in tertiary structure and conservation of active site peptide sequence. The conservation of protein structure suggests the common ancestry of the cytoskeletal elements formed by actin, found in eukaryotes, and MreB, found in prokaryotes. Indeed, recent studies have found that MreB proteins polymerize to form filaments that are similar to actin microfilaments. It has been shown to form multilayer sheets comprising diagonally interwoven filaments in the presence of ATP or GTP.

FtsZ is a protein encoded by the ftsZ gene that assembles into a ring at the future site of bacterial cell division. FtsZ is a prokaryotic homologue of the eukaryotic protein tubulin. The initials FtsZ mean "Filamenting temperature-sensitive mutant Z." The hypothesis was that cell division mutants of E. coli would grow as filaments due to the inability of the daughter cells to separate from one another. FtsZ is found in almost all bacteria, many archaea, all chloroplasts and some mitochondria, where it is essential for cell division. FtsZ assembles the cytoskeletal scaffold of the Z ring that, along with additional proteins, constricts to divide the cell in two.
Phalloidin belongs to a class of toxins called phallotoxins, which are found in the death cap mushroom (Amanita phalloides). It is a rigid bicyclic heptapeptide that is lethal after a few days when injected into the bloodstream. The major symptom of phalloidin poisoning is acute hunger due to the destruction of liver cells. It functions by binding and stabilizing filamentous actin (F-actin) and effectively prevents the depolymerization of actin fibers. Due to its tight and selective binding to F-actin, derivatives of phalloidin containing fluorescent tags are used widely in microscopy to visualize F-actin in biomedical research.
Tubulin in molecular biology can refer either to the tubulin protein superfamily of globular proteins, or one of the member proteins of that superfamily. α- and β-tubulins polymerize into microtubules, a major component of the eukaryotic cytoskeleton. It was discovered and named by Hideo Mōri in 1968. Microtubules function in many essential cellular processes, including mitosis. Tubulin-binding drugs kill cancerous cells by inhibiting microtubule dynamics, which are required for DNA segregation and therefore cell division.
Profilin is an actin-binding protein involved in the dynamic turnover and reconstruction of the actin cytoskeleton. It is found in most eukaryotic organisms. Profilin is important for spatially and temporally controlled growth of actin microfilaments, which is an essential process in cellular locomotion and cell shape changes. This restructuring of the actin cytoskeleton is essential for processes such as organ development, wound healing, and the hunting down of infectious intruders by cells of the immune system.
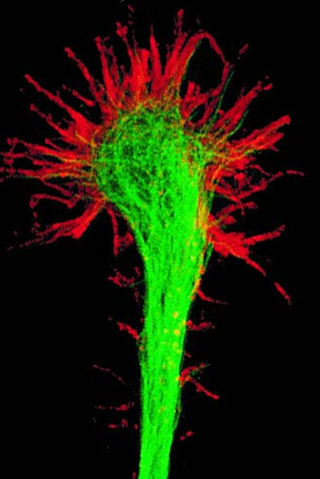
A growth cone is a large actin-supported extension of a developing or regenerating neurite seeking its synaptic target. It is the growth cone that drives axon growth. Their existence was originally proposed by Spanish histologist Santiago Ramón y Cajal based upon stationary images he observed under the microscope. He first described the growth cone based on fixed cells as "a concentration of protoplasm of conical form, endowed with amoeboid movements". Growth cones are situated on the tips of neurites, either dendrites or axons, of the nerve cell. The sensory, motor, integrative, and adaptive functions of growing axons and dendrites are all contained within this specialized structure.
ADF/cofilin is a family of actin-binding proteins associated with the rapid depolymerization of actin microfilaments that give actin its characteristic dynamic instability. This dynamic instability is central to actin's role in muscle contraction, cell motility and transcription regulation.
Cytochalasins are fungal metabolites that have the ability to bind to actin filaments and block polymerization and the elongation of actin. As a result of the inhibition of actin polymerization, cytochalasins can change cellular morphology, inhibit cellular processes such as cell division, and even cause cells to undergo apoptosis. Cytochalasins have the ability to permeate cell membranes, prevent cellular translocation and cause cells to enucleate. Cytochalasins can also have an effect on other aspects of biological processes unrelated to actin polymerization. For example, cytochalasin A and cytochalasin B can also inhibit the transport of monosaccharides across the cell membrane, cytochalasin H has been found to regulate plant growth, cytochalasin D inhibits protein synthesis and cytochalasin E prevents angiogenesis.
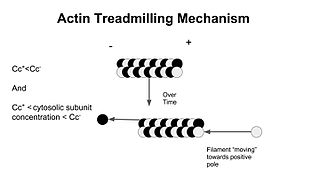
In molecular biology, treadmilling is a phenomenon observed within protein filaments of the cytoskeletons of many cells, especially in actin filaments and microtubules. It occurs when one end of a filament grows in length while the other end shrinks, resulting in a section of filament seemingly "moving" across a stratum or the cytosol. This is due to the constant removal of the protein subunits from these filaments at one end of the filament, while protein subunits are constantly added at the other end. Treadmilling was discovered by Wegner, who defined the thermodynamic and kinetic constraints. Wegner recognized that: “The equilibrium constant (K) for association of a monomer with a polymer is the same at both ends, since the addition of a monomer to each end leads to the same polymer.”; a simple reversible polymer can’t treadmill; ATP hydrolysis is required. GTP is hydrolyzed for microtubule treadmilling.

In biology, a protein filament is a long chain of protein monomers, such as those found in hair, muscle, or in flagella. Protein filaments form together to make the cytoskeleton of the cell. They are often bundled together to provide support, strength, and rigidity to the cell. When the filaments are packed up together, they are able to form three different cellular parts. The three major classes of protein filaments that make up the cytoskeleton include: actin filaments, microtubules and intermediate filaments.
Tropomodulin (TMOD) is a protein which binds and caps the minus end of actin, regulating the length of actin filaments in muscle and non-muscle cells.
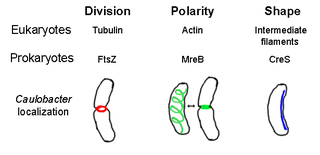
The prokaryotic cytoskeleton is the collective name for all structural filaments in prokaryotes. It was once thought that prokaryotic cells did not possess cytoskeletons, but advances in visualization technology and structure determination led to the discovery of filaments in these cells in the early 1990s. Not only have analogues for all major cytoskeletal proteins in eukaryotes been found in prokaryotes, cytoskeletal proteins with no known eukaryotic homologues have also been discovered. Cytoskeletal elements play essential roles in cell division, protection, shape determination, and polarity determination in various prokaryotes.
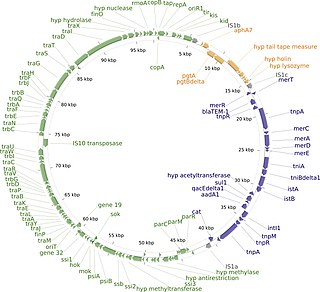
The R1 plasmid is a plasmid that was first isolated from Salmonella paratyphi bacteria in 1963. It is a short plasmid, composed of 97,566 nucleotides and 120 genes, that belongs to the IncFII plasmid group.
Actin remodeling is the biochemical process that allows for the dynamic alterations of cellular organization. The remodeling of actin filaments occurs in a cyclic pattern on cell surfaces and exists as a fundamental aspect to cellular life. During the remodeling process, actin monomers polymerize in response to signaling cascades that stem from environmental cues. The cell's signaling pathways cause actin to affect intracellular organization of the cytoskeleton and often consequently, the cell membrane. Again triggered by environmental conditions, actin filaments break back down into monomers and the cycle is completed. Actin-binding proteins (ABPs) aid in the transformation of actin filaments throughout the actin remodeling process. These proteins account for the diverse structure and changes in shape of Eukaryotic cells. Despite its complexity, actin remodeling may result in complete cytoskeletal reorganization in under a minute.
mDia1 is a member of the protein family called the formins and is a Rho effector. It is the mouse version of the diaphanous homolog 1 of Drosophila. mDia1 localizes to cells' mitotic spindle and midbody, plays a role in stress fiber and filopodia formation, phagocytosis, activation of serum response factor, formation of adherens junctions, and it can act as a transcription factor. mDia1 accelerates actin nucleation and elongation by interacting with barbed ends of actin filaments. The gene encoding mDia1 is located on Chromosome 18 of Mus musculus and named Diap1.
A plasmid partition system is a mechanism that ensures the stable inheritance of plasmids during bacterial cell division. Each plasmid has its independent replication system which controls the number of copies of the plasmid in a cell. The higher the copy number, the more likely the two daughter cells will contain the plasmid. Generally, each molecule of plasmid diffuses randomly, so the probability of having a plasmid-less daughter cell is 21−N, where N is the number of copies. For instance, if there are 2 copies of a plasmid in a cell, there is 50% chance of having one plasmid-less daughter cell. However, high-copy number plasmids have a cost for the hosting cell. This metabolic burden is lower for low-copy plasmids, but those have a higher probability of plasmid loss after a few generations. To control vertical transmission of plasmids, in addition to controlled-replication systems, bacterial plasmids use different maintenance strategies, such as multimer resolution systems, post-segregational killing systems, and partition systems.
References
- ↑ Gunning PW, Ghoshdastider U, Whitaker S, Popp D, Robinson RC (June 2015). "The evolution of compositionally and functionally distinct actin filaments". Journal of Cell Science. 128 (11): 2009–19. doi: 10.1242/jcs.165563 . PMID 25788699.
- ↑ Hoischen C, Bussiek M, Langowski J, Diekmann S (February 2008). "Escherichia coli low-copy-number plasmid R1 centromere parC forms a U-shaped complex with its binding protein ParR". Nucleic Acids Research. 36 (2): 607–15. doi:10.1093/nar/gkm672. PMC 2241845 . PMID 18056157.
- ↑ Popp D, Xu W, Narita A, Brzoska AJ, Skurray RA, Firth N, et al. (March 2010). "Structure and filament dynamics of the pSK41 actin-like ParM protein: implications for plasmid DNA segregation". The Journal of Biological Chemistry. 285 (13): 10130–40. doi: 10.1074/jbc.M109.071613 . PMC 2843175 . PMID 20106979.
- ↑ Popp D, Narita A, Ghoshdastider U, Maeda K, Maéda Y, Oda T, et al. (April 2010). "Polymeric structures and dynamic properties of the bacterial actin AlfA". Journal of Molecular Biology. 397 (4): 1031–41. doi:10.1016/j.jmb.2010.02.010. PMID 20156449.
- ↑ Popp D, Narita A, Lee LJ, Ghoshdastider U, Xue B, Srinivasan R, et al. (June 2012). "Novel actin-like filament structure from Clostridium tetani". The Journal of Biological Chemistry. 287 (25): 21121–9. doi: 10.1074/jbc.M112.341016 . PMC 3375535 . PMID 22514279.
- 1 2 3 Popp D, Narita A, Oda T, Fujisawa T, Matsuo H, Nitanai Y, et al. (February 2008). "Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability". The EMBO Journal. 27 (3): 570–9. doi:10.1038/sj.emboj.7601978. PMC 2241650 . PMID 18188150.
- 1 2 Garner EC, Campbell CS, Weibel DB, Mullins RD (March 2007). "Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog". Science. 315 (5816): 1270–4. Bibcode:2007Sci...315.1270G. doi:10.1126/science.1138527. PMC 2851738 . PMID 17332412.
- ↑ Garner EC, Campbell CS, Mullins RD (November 2004). "Dynamic instability in a DNA-segregating prokaryotic actin homolog". Science. 306 (5698): 1021–5. Bibcode:2004Sci...306.1021G. doi:10.1126/science.1101313. PMID 15528442. S2CID 14032209.