
Intermediate filaments (IFs) are cytoskeletal structural components found in the cells of vertebrates, and many invertebrates. Homologues of the IF protein have been noted in an invertebrate, the cephalochordate Branchiostoma.

A desmosome, also known as a macula adherens, is a cell structure specialized for cell-to-cell adhesion. A type of junctional complex, they are localized spot-like adhesions randomly arranged on the lateral sides of plasma membranes. Desmosomes are one of the stronger cell-to-cell adhesion types and are found in tissue that experience intense mechanical stress, such as cardiac muscle tissue, bladder tissue, gastrointestinal mucosa, and epithelia.

Arrhythmogenic cardiomyopathy (ACM), arrhythmogenic right ventricular dysplasia (ARVD), or arrhythmogenic right ventricular cardiomyopathy (ARVC), most commonly is an inherited heart disease.

Desmin is a protein that in humans is encoded by the DES gene. Desmin is a muscle-specific, type III intermediate filament that integrates the sarcolemma, Z disk, and nuclear membrane in sarcomeres and regulates sarcomere architecture.

Desmoglein-1 is a protein that in humans is encoded by the DSG1 gene. Desmoglein-1 is expressed everywhere in the skin epidermis, but mainly it is expressed in the superficial upper layers of the skin epidermis.
Desmoglein-2 is a protein that in humans is encoded by the DSG2 gene. Desmoglein-2 is highly expressed in epithelial cells and cardiomyocytes. Desmoglein-2 is localized to desmosome structures at regions of cell-cell contact and functions to structurally adhere adjacent cells together. In cardiac muscle, these regions are specialized regions known as intercalated discs. Mutations in desmoglein-2 have been associated with arrhythmogenic right ventricular cardiomyopathy and familial dilated cardiomyopathy.

Desmoplakin is a protein in humans that is encoded by the DSP gene. Desmoplakin is a critical component of desmosome structures in cardiac muscle and epidermal cells, which function to maintain the structural integrity at adjacent cell contacts. In cardiac muscle, desmoplakin is localized to intercalated discs which mechanically couple cardiac cells to function in a coordinated syncytial structure. Mutations in desmoplakin have been shown to play a role in dilated cardiomyopathy and arrhythmogenic right ventricular cardiomyopathy, where it may present with acute myocardial injury; striate palmoplantar keratoderma, Carvajal syndrome and paraneoplastic pemphigus.

Plakoglobin, also known as junction plakoglobin or gamma-catenin, is a protein that in humans is encoded by the JUP gene. Plakoglobin is a member of the catenin protein family and homologous to β-catenin. Plakoglobin is a cytoplasmic component of desmosomes and adherens junctions structures located within intercalated discs of cardiac muscle that function to anchor sarcomeres and join adjacent cells in cardiac muscle. Mutations in plakoglobin are associated with arrhythmogenic right ventricular dysplasia.
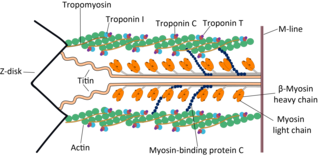
Cardiac muscle troponin T (cTnT) is a protein that in humans is encoded by the TNNT2 gene. Cardiac TnT is the tropomyosin-binding subunit of the troponin complex, which is located on the thin filament of striated muscles and regulates muscle contraction in response to alterations in intracellular calcium ion concentration.

Ryanodine receptor 2 (RYR2) is one of a class of ryanodine receptors and a protein found primarily in cardiac muscle. In humans, it is encoded by the RYR2 gene. In the process of cardiac calcium-induced calcium release, RYR2 is the major mediator for sarcoplasmic release of stored calcium ions.

The myosin-binding protein C, cardiac-type is a protein that in humans is encoded by the MYBPC3 gene. This isoform is expressed exclusively in heart muscle during human and mouse development, and is distinct from those expressed in slow skeletal muscle (MYBPC1) and fast skeletal muscle (MYBPC2).

Desmocollin-2 is a protein that in humans is encoded by the DSC2 gene. Desmocollin-2 is a cadherin-type protein that functions to link adjacent cells together in specialized regions known as desmosomes. Desmocollin-2 is widely expressed, and is the only desmocollin isoform expressed in cardiac muscle, where it localizes to intercalated discs. Mutations in DSC2 have been causally linked to arrhythmogenic right ventricular cardiomyopathy.

Plakophilin-1 is a protein that in humans is encoded by the PKP1 gene.

Desmocollin-3 is a protein that in humans is encoded by the DSC3 gene.

Plakophilin-3 is a protein that in humans is encoded by the PKP3 gene.

LIM domain binding 3 (LDB3), also known as Z-band alternatively spliced PDZ-motif (ZASP), is a protein which in humans is encoded by the LDB3 gene. ZASP belongs to the Enigma subfamily of proteins and stabilizes the sarcomere during contraction, through interactions with actin in cardiac and skeletal muscles. Mutations in the ZASP gene has been associated with several muscular diseases.

Transmembrane protein 43 is a protein that in humans is encoded by the TMEM43 gene. TMEM43 may have an important role in maintaining nuclear envelope structure by organizing protein complexes at the inner nuclear membrane. Required for retaining emerin at the inner nuclear membrane. However, the localization of TMEM43 in myocardial tissue is controversial discussed. Franke et al. demonstrated that TMEM43 is localized at the intercalated disc but not at the nuclear envelope. In contrast Christensen et al. have shown that TMEM43 is mainly localized at the sarcolemma. Mutations in TMEM43 are associated with ARVD and EDMD7.

Ankyrin-2, also known as Ankyrin-B, and Brain ankyrin, is a protein which in humans is encoded by the ANK2 gene. Ankyrin-2 is ubiquitously expressed, but shows high expression in cardiac muscle. Ankyrin-2 plays an essential role in the localization and membrane stabilization of ion transporters and ion channels in cardiomyocytes, as well as in costamere structures. Mutations in ANK2 cause a dominantly-inherited, cardiac arrhythmia syndrome known as long QT syndrome 4 as well as sick sinus syndrome; mutations have also been associated to a lesser degree with hypertrophic cardiomyopathy. Alterations in ankyrin-2 expression levels are observed in human heart failure.
Desmocollins are a subfamily of desmosomal cadherins, the transmembrane constituents of desmosomes. They are co-expressed with desmogleins to link adjacent cells by extracellular adhesion. There are seven desmosomal cadherins in humans, three desmocollins and four desmogleins. Desmosomal cadherins allow desmosomes to contribute to the integrity of tissue structure in multicellular living organisms.
Frank I. Marcus was an American cardiologist and Emeritus Professor of Medicine at the University of Arizona Health Sciences Center, the author of more than 290 publications in peer-reviewed medical journals and of 90 book chapters. He was considered a world expert on arrhythmogenic right ventricular cardiomyopathy (ARVC) and was a member of the Editorial/Scientific Board of 14 Cardiovascular Journals as well as a reviewer for 26 other medical publications.




















