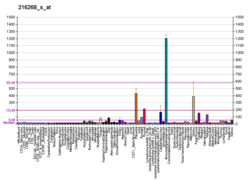
The Notch signaling pathway is a highly conserved cell signaling system present in most animals. Mammals possess four different notch receptors, referred to as NOTCH1, NOTCH2, NOTCH3, and NOTCH4. The notch receptor is a single-pass transmembrane receptor protein. It is a hetero-oligomer composed of a large extracellular portion, which associates in a calcium-dependent, non-covalent interaction with a smaller piece of the notch protein composed of a short extracellular region, a single transmembrane-pass, and a small intracellular region.

Alagille syndrome (ALGS) is a genetic disorder that affects primarily the liver and the heart. Problems associated with the disorder generally become evident in infancy or early childhood. The disorder is inherited in an autosomal dominant pattern, and the estimated prevalence of Alagille syndrome is 1 in every 30,000 to 1 in every 40,000 live births. It is named after the French pediatrician Daniel Alagille, who first described the condition in 1969.

Hajdu–Cheney syndrome, also called acroosteolysis with osteoporosis and changes in skull and mandible, arthrodentoosteodysplasia and Cheney syndrome, is an extremely rare autosomal dominant congenital disorder of the connective tissue characterized by severe and excessive bone resorption leading to osteoporosis and a wide range of other possible symptoms. Mutations in the NOTCH2 gene, identified in 2011, cause HCS. HCS is so rare that only about 50 cases have been reported worldwide since the discovery of the syndrome in 1948

Caveolin-3 is a protein that in humans is encoded by the CAV3 gene. Alternative splicing has been identified for this locus, with inclusion or exclusion of a differentially spliced intron. In addition, transcripts utilize multiple polyA sites and contain two potential translation initiation sites.

Neurogenic locus notch homolog protein 3(Notch 3) is a protein that in humans is encoded by the NOTCH3 gene.

The photoreceptor cell-specific nuclear receptor (PNR), also known as NR2E3, is a protein that in humans is encoded by the NR2E3 gene. PNR is a member of the nuclear receptor super family of intracellular transcription factors.

Neurogenic locus notch homolog protein 1(Notch 1) is a protein encoded in humans by the NOTCH1 gene. Notch 1 is a single-pass transmembrane receptor.
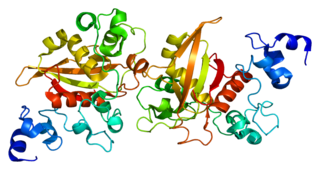
Neurogenic locus notch homolog protein 2 is a protein that in humans is encoded by the NOTCH2 gene.

G-protein coupled receptor 143, also known as Ocular albinism type 1 (OA1) in humans, is a conserved integral membrane protein with seven transmembrane domains and similarities with G protein-coupled receptors (GPCRs) that is expressed in the eye and epidermal melanocytes. This protein encoded by the GPR143 gene, whose variants can lead to Ocular albinism type 1.

Delta-like protein 1 is a protein that in humans is encoded by the DLL1 gene.

Recombination signal binding protein for immunoglobulin kappa J region is a protein that in humans is encoded by the RBPJ gene.

Endothelin-3 is a protein that in humans is encoded by the EDN3 gene.

Hairy/enhancer-of-split related with YRPW motif protein 2 (HEY2) also known as cardiovascular helix-loop-helix factor 1 (CHF1) is a protein that in humans is encoded by the HEY2 gene.

Jagged-2 is a protein that in humans is encoded by the JAG2 gene.

Beta-1,3-N-acetylglucosaminyltransferase manic fringe is an enzyme that in humans is encoded by the MFNG gene, a member of the fringe gene family which also includes the radical fringe (RFNG) and lunatic fringe (LFNG).

Acetylcholine receptor subunit delta is a protein that in humans is encoded by the CHRND gene.

Cytokine receptor-like factor 1 is a protein that in humans is encoded by the CRLF1 gene.

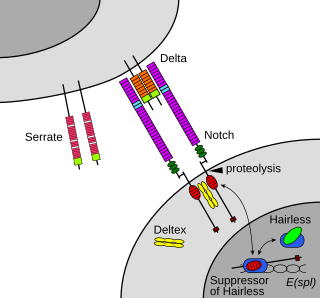
Notch proteins are a family of type 1 transmembrane proteins that form a core component of the Notch signaling pathway, which is highly conserved in animals. The Notch extracellular domain mediates interactions with DSL family ligands, allowing it to participate in juxtacrine signaling. The Notch intracellular domain acts as a transcriptional activator when in complex with CSL family transcription factors. Members of this type 1 transmembrane protein family share several core structures, including an extracellular domain consisting of multiple epidermal growth factor (EGF)-like repeats and an intracellular domain transcriptional activation domain (TAD). Notch family members operate in a variety of different tissues and play a role in a variety of developmental processes by controlling cell fate decisions. Much of what is known about Notch function comes from studies done in Caenorhabditis elegans (C.elegans) and Drosophila melanogaster. Human homologs have also been identified, but details of Notch function and interactions with its ligands are not well known in this context.

Transmembrane protein 216 is a protein in humans that is encoded by the TMEM216 gene.
In molecular biology, there are a number of neurogenic proteins referred to as mastermind-like proteins (MAMLs) of which this domain is the N-terminal region. Mastermind-like proteins act as critical transcriptional co-activators for Notch signaling.