COVID-20, COVID-21 or COVID-22 may refer to:
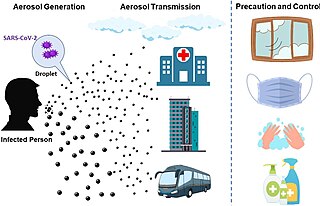
The transmission of COVID-19 is the passing of coronavirus disease 2019 from person to person. COVID-19 is mainly transmitted when people breathe in air contaminated by droplets/aerosols and small airborne particles containing the virus. Infected people exhale those particles as they breathe, talk, cough, sneeze, or sing. Transmission is more likely the closer people are. However, infection can occur over longer distances, particularly indoors.

Both the American mink and the European mink have shown high susceptibility to SARS-CoV-2 since the earliest stages of the COVID-19 pandemic, first in mink farms across Europe, followed by mink farms in the United States. Mortality has been extremely high among mink, with 35–55% of infected adult animals dying from COVID-19 in a study of farmed mink in the U.S. state of Utah.

The Alpha variant (B.1.1.7) was a SARS-CoV-2 variant of concern. It was estimated to be 40–80% more transmissible than the wild-type SARS-CoV-2. Scientists more widely took note of this variant in early December 2020, when a phylogenetic tree showing viral sequences from Kent, United Kingdom looked unusual.

The Beta variant, (B.1.351), was a variant of SARS-CoV-2, the virus that causes COVID-19. One of several SARS-CoV-2 variants initially believed to be of particular importance, it was first detected in the Nelson Mandela Bay metropolitan area of the Eastern Cape province of South Africa in October 2020, which was reported by the country's health department on 18 December 2020. Phylogeographic analysis suggests this variant emerged in the Nelson Mandela Bay area in July or August 2020.

Variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are viruses that, while similar to the original, have genetic changes that are of enough significance to lead virologists to label them separately. SARS-CoV-2 is the virus that causes coronavirus disease 2019 (COVID-19). Some have been stated, to be of particular importance due to their potential for increased transmissibility, increased virulence, or reduced effectiveness of vaccines against them. These variants contribute to the continuation of the COVID-19 pandemic.

The Gamma variant (P.1) was one of the variants of SARS-CoV-2, the virus that causes COVID-19. This variant of SARS-CoV-2 has been named lineage P.1 and has 17 amino acid substitutions, ten of which in its spike protein, including these three designated to be of particular concern: N501Y, E484K and K417T. It was first detected by the National Institute of Infectious Diseases (NIID) of Japan, on 6 January 2021 in four people who had arrived in Tokyo having visited Amazonas, Brazil, four days earlier. It was subsequently declared to be in circulation in Brazil. Under the simplified naming scheme proposed by the World Health Organization, P.1 was labeled Gamma variant, and was considered a variant of concern until March 2022, when it was largely displaced by the delta and omicron variants.

Iota variant, also known as lineage B.1.526, is one of the variants of SARS-CoV-2, the virus that causes COVID-19. It was first detected in New York City in November 2020. The variant has appeared with two notable mutations: the E484K spike mutation, which may help the virus evade antibodies, and the S477N mutation, which helps the virus bind more tightly to human cells.

The Delta variant (B.1.617.2) was a variant of SARS-CoV-2, the virus that causes COVID-19. It was first detected in India on 5 October 2020. The Delta variant was named on 31 May 2021 and had spread to over 179 countries by 22 November 2021. The World Health Organization (WHO) indicated in June 2021 that the Delta variant was becoming the dominant strain globally.

Theta variant, also known as lineage P.3, is one of the variants of SARS-CoV-2, the virus that causes COVID-19. The variant was first identified in the Philippines on February 18, 2021, when two mutations of concern were detected in Central Visayas. It was detected in Japan on March 12, 2021, when a traveler from the Philippines arrived at Narita International Airport in Tokyo.

Kappa variant is a variant of SARS-CoV-2, the virus that causes COVID-19. It is one of the three sublineages of Pango lineage B.1.617. The SARS-CoV-2 Kappa variant is also known as lineage B.1.617.1 and was first detected in India in December 2020. By the end of March 2021, the Kappa sub-variant accounted for more than half of the sequences being submitted from India. On 1 April 2021, it was designated a Variant Under Investigation (VUI-21APR-01) by Public Health England.
Lineage B.1.617 is a lineage of SARS-CoV-2, the virus that causes COVID-19. It first came to international attention in late March 2021 after the newly established INSACOG performed genome sequencing on positive samples throughout various Indian states. Analysis of samples from Maharashtra had revealed that compared to December 2020, there was an increase in the fraction of samples with the E484Q and L452R mutations. Lineage B.1.617 later came to be dubbed a double mutant by news media.

The Lambda variant, also known as lineage C.37, is a variant of SARS-CoV-2, the virus that causes COVID-19. It was first detected in Peru in August 2020. On 14 June 2021, the World Health Organization (WHO) named it Lambda variant and designated it as a variant of interest. On 16 March 2022, the WHO has de-escalated the Lambda variant to "previously circulating variants of concern".

Epsilon variant, also known as CAL.20C and referring to two PANGO lineages B.1.427 and B.1.429, is one of the variants of SARS-CoV-2, the virus that causes COVID-19. It was first detected in California, USA in July 2020.

Zeta variant, also known as lineage P.2, is a variant of SARS-CoV-2, the virus that causes COVID-19. It was first detected in the state of Rio de Janeiro; it harbors the E484K mutation, but not the N501Y and K417T mutations. It evolved independently in Rio de Janeiro without being directly related to the Gamma variant from Manaus.

The Eta variant is a variant of SARS-CoV-2, the virus that causes COVID-19. The Eta variant or lineage B.1.525, also called VUI-21FEB-03 by Public Health England (PHE) and formerly known as UK1188, 21D or 20A/S:484K, does not carry the same N501Y mutation found in Alpha, Beta and Gamma, but carries the same E484K-mutation as found in the Gamma, Zeta, and Beta variants, and also carries the same ΔH69/ΔV70 deletion as found in Alpha, N439K variant and Y453F variant.

The Mu variant, also known as lineage B.1.621 or VUI-21JUL-1, is one of the variants of SARS-CoV-2, the virus that causes COVID-19. It was first detected in Colombia in January 2021 and was designated by the WHO as a variant of interest on August 30, 2021. On 16 March 2022, the WHO has de-escalated the Mu variant and its subvariants to "previously circulating variants of concern".

Omicron (B.1.1.529) is a variant of SARS-CoV-2 first reported to the World Health Organization (WHO) by the Network for Genomics Surveillance in South Africa on 24 November 2021. It was first detected in Botswana and has spread to become the predominant variant in circulation around the world. Following the original B.1.1.529 variant, several subvariants of Omicron have emerged including: BA.1, BA.2, BA.3, BA.4, and BA.5. Since October 2022, two subvariants of BA.5 called BQ.1 and BQ.1.1 have emerged.

This timeline of the SARS-CoV-2 Omicron variant is a dynamic list, and as such may never satisfy criteria of completeness. Some events may only be fully understood and/or discovered in retrospect.

Blood samples gathered by USDA researchers in 2021 showed that 40% of sampled white-tailed deer demonstrated evidence of SARS-CoV-2 antibodies, with the highest percentages in Michigan, at 67%, and Pennsylvania, at 44%. A later study by Penn State University and wildlife officials in Iowa showed that up to 80% of Iowa deer sampled from April 2020 through January 2021 had tested positive for active SARS-CoV-2 infection, rather than solely antibodies from prior infection. This data, confirmed by the National Veterinary Services Laboratory, alerted scientists to the possibility that white-tailed deer had become a natural reservoir for the coronavirus, serving as a potential "variant factory" for eventual retransmission back into humans.





