| Part of a series on the |
| COVID-19 pandemic |
|---|
 |
|
| |
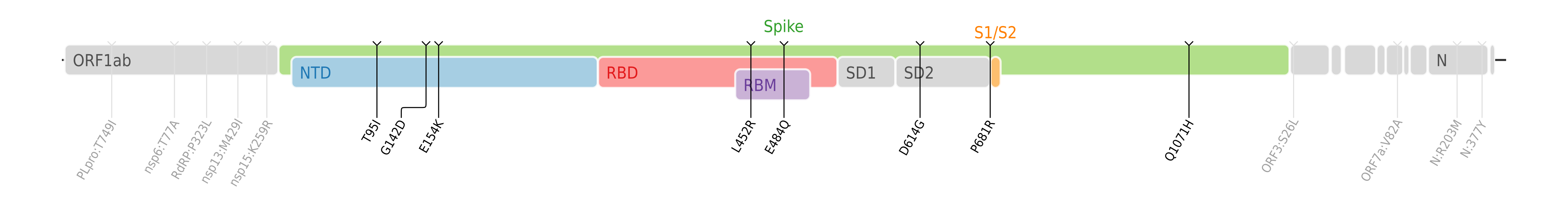
Kappa variant [1] is a variant of SARS-CoV-2, the virus that causes COVID-19. It is one of the three sublineages of Pango lineage B.1.617. The SARS-CoV-2 Kappa variant is also known as lineage B.1.617.1 and was first detected in India in December 2020. [2] By the end of March 2021, the Kappa sub-variant accounted for more than half of the sequences being submitted from India. [3] On 1 April 2021, it was designated a Variant Under Investigation (VUI-21APR-01) by Public Health England. [4]