Arabidopsis thaliana, the thale cress, mouse-ear cress or arabidopsis, is a small plant from the mustard family (Brassicaceae), native to Eurasia and Africa. Commonly found along the shoulders of roads and in disturbed land, it is generally considered a weed.
Gibberellins (GAs) are plant hormones that regulate various developmental processes, including stem elongation, germination, dormancy, flowering, flower development, and leaf and fruit senescence. GAs are one of the longest-known classes of plant hormone. It is thought that the selective breeding of crop strains that were deficient in GA synthesis was one of the key drivers of the "green revolution" in the 1960s, a revolution that is credited to have saved over a billion lives worldwide.

Abscisic acid is a plant hormone. ABA functions in many plant developmental processes, including seed and bud dormancy, the control of organ size and stomatal closure. It is especially important for plants in the response to environmental stresses, including drought, soil salinity, cold tolerance, freezing tolerance, heat stress and heavy metal ion tolerance.

In evolutionary developmental biology, homeosis is the transformation of one organ into another, arising from mutation in or misexpression of certain developmentally critical genes, specifically homeotic genes. In animals, these developmental genes specifically control the development of organs on their anteroposterior axis. In plants, however, the developmental genes affected by homeosis may control anything from the development of a stamen or petals to the development of chlorophyll. Homeosis may be caused by mutations in Hox genes, found in animals, or others such as the MADS-box family in plants. Homeosis is a characteristic that has helped insects become as successful and diverse as they are.
In molecular biology, a CCAAT box is a distinct pattern of nucleotides with GGCCAATCT consensus sequence that occur upstream by 60–100 bases to the initial transcription site. The CAAT box signals the binding site for the RNA transcription factor, and is typically accompanied by a conserved consensus sequence. It is an invariant DNA sequence at about minus 70 base pairs from the origin of transcription in many eukaryotic promoters. Genes that have this element seem to require it for the gene to be transcribed in sufficient quantities. It is frequently absent from genes that encode proteins used in virtually all cells. This box along with the GC box is known for binding general transcription factors. Both of these consensus sequences belong to the regulatory promoter. Full gene expression occurs when transcription activator proteins bind to each module within the regulatory promoter. Protein specific binding is required for the CCAAT box activation. These proteins are known as CCAAT box binding proteins/CCAAT box binding factors.
Cryptochromes are a class of flavoproteins found in plants and animals that are sensitive to blue light. They are involved in the circadian rhythms and the sensing of magnetic fields in a number of species. The name cryptochrome was proposed as a portmanteau combining the chromatic nature of the photoreceptor, and the cryptogamic organisms on which many blue-light studies were carried out.
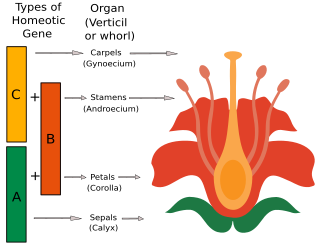
The ABC model of flower development is a scientific model of the process by which flowering plants produce a pattern of gene expression in meristems that leads to the appearance of an organ oriented towards sexual reproduction, a flower. There are three physiological developments that must occur in order for this to take place: firstly, the plant must pass from sexual immaturity into a sexually mature state ; secondly, the transformation of the apical meristem's function from a vegetative meristem into a floral meristem or inflorescence; and finally the growth of the flower's individual organs. The latter phase has been modelled using the ABC model, which aims to describe the biological basis of the process from the perspective of molecular and developmental genetics.
The MADS box is a conserved sequence motif. The genes which contain this motif are called the MADS-box gene family. The MADS box encodes the DNA-binding MADS domain. The MADS domain binds to DNA sequences of high similarity to the motif CC[A/T]6GG termed the CArG-box. MADS-domain proteins are generally transcription factors. The length of the MADS-box reported by various researchers varies somewhat, but typical lengths are in the range of 168 to 180 base pairs, i.e. the encoded MADS domain has a length of 56 to 60 amino acids. There is evidence that the MADS domain evolved from a sequence stretch of a type II topoisomerase in a common ancestor of all extant eukaryotes.
Superman is a plant gene in Arabidopsis thaliana, that plays a role in controlling the boundary between stamen and carpel development in a flower. It is named for the comic book character Superman, and the related genes kryptonite (gene) and clark kent were named accordingly. It encodes a transcription factor. Homologous genes are known in the petunia and snapdragon, which are also involved in flower development, although in both cases there are important differences from the functioning in Arabidopsis. Superman is expressed early on in flower development, in the stamen whorl adjacent to the carpel whorl. It interacts with the other genes of the ABC model of flower development in a variety of ways.

Myb genes are part of a large gene family of transcription factors found in animals and plants. In humans, it includes Myb proto-oncogene like 1 and Myb-related protein B in addition to MYB proper. Members of the extended SANT/Myb family also include the SANT domain and other similar all-helical homeobox-like domains.

D site of albumin promoter binding protein, also known as DBP, is a protein which in humans is encoded by the DBP gene.
The B3 DNA binding domain (DBD) is a highly conserved domain found exclusively in transcription factors combined with other domains. It consists of 100-120 residues, includes seven beta strands and two alpha helices that form a DNA-binding pseudobarrel protein fold ; it interacts with the major groove of DNA.
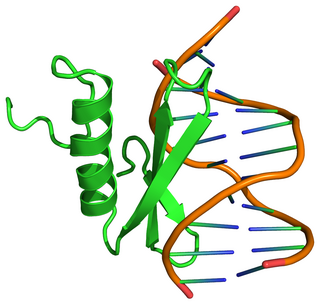
Ethylene-responsive element binding protein(EREBP) is a homeobox gene from Arabidopsis thaliana and other plants which encodes a transcription factor. EREBP is responsible in part for mediating the response in plants to the plant hormone ethylene.
Agamous (AG) is a homeotic gene and MADS-box transcription factor from Arabidopsis thaliana. The TAIR AGI number is AT4G18960.

The WRKY domain is found in the WRKY transcription factor family, a class of transcription factors. The WRKY domain is found almost exclusively in plants although WRKY genes appear present in some diplomonads, social amoebae and other amoebozoa, and fungi incertae sedis. They appear absent in other non-plant species. WRKY transcription factors have been a significant area of plant research for the past 20 years. The WRKY DNA-binding domain recognizes the W-box (T)TGAC(C/T) cis-regulatory element.
Timing of CAB expression 1 is a protein that in Arabidopsis thaliana is encoded by the TOC1 gene. TOC1 is also known as two-component response regulator-like APRR1.
WRKY transcription factors are proteins that bind DNA. They are transcription factors that regulate many processes in plants and algae (Viridiplantae), such as the responses to biotic and abiotic stresses, senescence, seed dormancy and seed germination and some developmental processes but also contribute to secondary metabolism.
Circadian Clock Associated 1 (CCA1) is a gene that is central to the circadian oscillator of angiosperms. It was first identified in Arabidopsis thaliana in 1993. CCA1 interacts with LHY and TOC1 to form the core of the oscillator system. CCA1 expression peaks at dawn. Loss of CCA1 function leads to a shortened period in the expression of many other genes.
LUX or Phytoclock1 (PCL1) is a gene that codes for LUX ARRHYTHMO, a protein necessary for circadian rhythms in Arabidopsis thaliana. LUX protein associates with Early Flowering 3 (ELF3) and Early Flowering 4 (ELF4) to form the Evening Complex (EC), a core component of the Arabidopsis repressilator model of the plant circadian clock. The LUX protein functions as a transcription factor that negatively regulates Pseudo-Response Regulator 9 (PRR9), a core gene of the Midday Complex, another component of the Arabidopsis repressilator model. LUX is also associated with circadian control of hypocotyl growth factor genes PHYTOCHROME INTERACTING FACTOR 4 (PIF4) and PHYTOCHROME INTERACTING FACTOR 5 (PIF5).
The chlorophyll a/b-binding protein gene, otherwise known as the CAB gene, is one of the most thoroughly characterized clock-regulated genes in plants. There are a variety of CAB proteins that are derived from this gene family. Studies on Arabidopsis plants have shed light on the mechanisms of biological clocks under the regulation of CAB genes. Dr. Steve Kay discovered that CAB was regulated by a circadian clock, which switched the gene on in the morning and off in the late afternoon. The genes code for proteins that associate with chlorophyll and xanthophylls. This association aids the absorption of sunlight, which transfers energy to photosystem II to drive photosynthetic electron transport.