CLOCK is a gene encoding a basic helix-loop-helix-PAS transcription factor that is known to affect both the persistence and period of circadian rhythms.
Neuromedin B (NMB) is a bombesin-related peptide in mammals. It was originally purified from pig spinal cord, and later shown to be present in human central nervous system and gastrointestinal tract.
An E-box is a DNA response element found in some eukaryotes that acts as a protein-binding site and has been found to regulate gene expression in neurons, muscles, and other tissues. Its specific DNA sequence, CANNTG, with a palindromic canonical sequence of CACGTG, is recognized and bound by transcription factors to initiate gene transcription. Once the transcription factors bind to the promoters through the E-box, other enzymes can bind to the promoter and facilitate transcription from DNA to mRNA.
Period (per) is a gene located on the X chromosome of Drosophila melanogaster. Oscillations in levels of both per transcript and its corresponding protein PER have a period of approximately 24 hours and together play a central role in the molecular mechanism of the Drosophila biological clock driving circadian rhythms in eclosion and locomotor activity. Mutations in the per gene can shorten (perS), lengthen (perL), and even abolish (per0) the period of the circadian rhythm.

Period circadian protein homolog 1 is a protein in humans that is encoded by the PER1 gene.

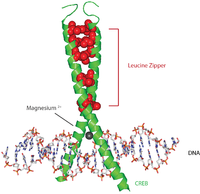
CAMP responsive element binding protein 1, also known as CREB-1, is a protein that in humans is encoded by the CREB1 gene. This protein binds the cAMP response element, a DNA nucleotide sequence present in many viral and cellular promoters. The binding of CREB1 stimulates transcription.

Cyclic AMP-dependent transcription factor ATF-1 is a protein that in humans is encoded by the ATF1 gene.

Activating transcription factor 4 , also known as ATF4, is a protein that in humans is encoded by the ATF4 gene.

Activating transcription factor 2, also known as ATF2, is a protein that, in humans, is encoded by the ATF2 gene.
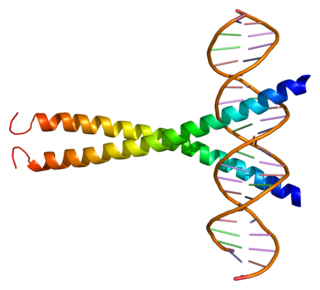
cAMP responsive element modulator is a protein that in humans is encoded by the CREM gene, and it belongs to the cAMP-responsive element binding protein family. It has multiple isoforms, which act either as repressors or activators. CREB family is important for in regulating transcription in response to various stresses, metabolic and developmental signals. CREM transcription factors also play an important role in many physiological systems, such as cardiac function, circadian rhythms, locomotion and spermatogenesis.

Ribosomal protein S6 kinase alpha-2 is an enzyme that in humans is encoded by the RPS6KA2 gene.

CREB-regulated transcription coactivator 1 (CRTC1), previously referred to as TORC1 (Transducer Of Regulated CREB activity 1), is a protein that in humans is encoded by the CRTC1 gene. It is expressed in a limited number of tissues that include fetal brain and liver and adult heart, skeletal muscles, liver and salivary glands and various regions of the adult central nervous system.

CREB regulated transcription coactivator 2, also known as CRTC2, is a protein which in humans is encoded by the CRTC2 gene.
In molecular biology, an oscillating gene is a gene that is expressed in a rhythmic pattern or in periodic cycles. Oscillating genes are usually circadian and can be identified by periodic changes in the state of an organism. Circadian rhythms, controlled by oscillating genes, have a period of approximately 24 hours. For example, plant leaves opening and closing at different times of the day or the sleep-wake schedule of animals can all include circadian rhythms. Other periods are also possible, such as 29.5 days resulting from circalunar rhythms or 12.4 hours resulting from circatidal rhythms. Oscillating genes include both core clock component genes and output genes. A core clock component gene is a gene necessary for to the pacemaker. However, an output oscillating gene, such as the AVP gene, is rhythmic but not necessary to the pacemaker.
Joseph S. Takahashi is a Japanese American neurobiologist and geneticist. Takahashi is a professor at University of Texas Southwestern Medical Center as well as an investigator at the Howard Hughes Medical Institute. Takahashi's research group discovered the genetic basis for the mammalian circadian clock in 1994 and identified the Clock gene in 1997. Takahashi was elected to the National Academy of Sciences in 2003.
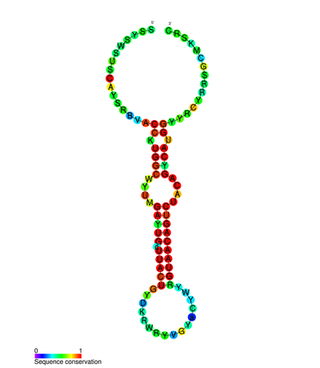
In molecular biology miR-132 microRNA is a short non-coding RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms, generally reducing protein levels through the cleavage of mRNAs or the repression of their translation. Several targets for miR-132 have been described, including mediators of neurological development, synaptic transmission, inflammation and angiogenesis.
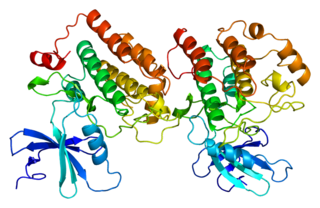
Casein kinase I isoform epsilon or CK1ε, is an enzyme that is encoded by the CSNK1E gene in humans. It is the mammalian homolog of doubletime. CK1ε is a serine/threonine protein kinase and is very highly conserved; therefore, this kinase is very similar to other members of the casein kinase 1 family, of which there are seven mammalian isoforms. CK1ε is most similar to CK1δ in structure and function as the two enzymes maintain a high sequence similarity on their regulatory C-terminal and catalytic domains. This gene is a major component of the mammalian oscillator which controls cellular circadian rhythms. CK1ε has also been implicated in modulating various human health issues such as cancer, neurodegenerative diseases, and diabetes.
Transcription-translation feedback loop (TTFL) is a cellular model for explaining circadian rhythms in behavior and physiology. Widely conserved across species, the TTFL is auto-regulatory, in which transcription of clock genes is regulated by their own protein products.
The food-entrainable oscillator (FEO) is a circadian clock that can be entrained by varying the time of food presentation. It was discovered when a rhythm was found in rat activity. This was called food anticipatory activity (FAA), and this is when the wheel-running activity of mice decreases after feeding, and then rapidly increases in the hours leading up to feeding. FAA appears to be present in non-mammals (pigeons/fish), but research heavily focuses on its presence in mammals. This rhythmic activity does not require the suprachiasmatic nucleus (SCN), the central circadian oscillator in mammals, implying the existence of an oscillator, the FEO, outside of the SCN, but the mechanism and location of the FEO is not yet known. There is ongoing research to investigate if the FEO is the only non-light entrainable oscillator in the body.

The arginine vasopressin gene (AVP) is a gene whose product is proteolytically cleaved to produce vasopressin, neurophysin II, and a glycoprotein called copeptin. AVP and other AVP-like peptides are found in mammals, as well as mollusks, arthropods, nematodes, and other invertebrate species. In humans, AVP is present on chromosome 20 and plays a role in homeostatic regulation. The products of AVP have many functions that include vasoconstriction, regulating the balance of water in the body, and regulating responses to stress. Expression of AVP is regulated by the transcription translation feedback loop (TTFL), which is an important part of the circadian system that controls the expression of clock genes. AVP has important implications in the medical field as its products have significant roles throughout body.