Gluconeogenesis (GNG) is a metabolic pathway that results in the biosynthesis of glucose from certain non-carbohydrate carbon substrates. It is a ubiquitous process, present in plants, animals, fungi, bacteria, and other microorganisms. In vertebrates, gluconeogenesis occurs mainly in the liver and, to a lesser extent, in the cortex of the kidneys. It is one of two primary mechanisms – the other being degradation of glycogen (glycogenolysis) – used by humans and many other animals to maintain blood sugar levels, avoiding low levels (hypoglycemia). In ruminants, because dietary carbohydrates tend to be metabolized by rumen organisms, gluconeogenesis occurs regardless of fasting, low-carbohydrate diets, exercise, etc. In many other animals, the process occurs during periods of fasting, starvation, low-carbohydrate diets, or intense exercise.
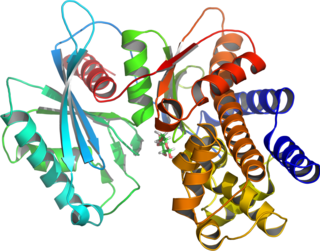
Glucokinase is an enzyme that facilitates phosphorylation of glucose to glucose-6-phosphate. Glucokinase occurs in cells in the liver and pancreas of humans and most other vertebrates. In each of these organs it plays an important role in the regulation of carbohydrate metabolism by acting as a glucose sensor, triggering shifts in metabolism or cell function in response to rising or falling levels of glucose, such as occur after a meal or when fasting. Mutations of the gene for this enzyme can cause unusual forms of diabetes or hypoglycemia.

The insulin receptor (IR) is a transmembrane receptor that is activated by insulin, IGF-I, IGF-II and belongs to the large class of receptor tyrosine kinase. Metabolically, the insulin receptor plays a key role in the regulation of glucose homeostasis; a functional process that under degenerate conditions may result in a range of clinical manifestations including diabetes and cancer. Insulin signalling controls access to blood glucose in body cells. When insulin falls, especially in those with high insulin sensitivity, body cells begin only to have access to lipids that do not require transport across the membrane. So, in this way, insulin is the key regulator of fat metabolism as well. Biochemically, the insulin receptor is encoded by a single gene INSR, from which alternate splicing during transcription results in either IR-A or IR-B isoforms. Downstream post-translational events of either isoform result in the formation of a proteolytically cleaved α and β subunit, which upon combination are ultimately capable of homo or hetero-dimerisation to produce the ≈320 kDa disulfide-linked transmembrane insulin receptor.
In biochemistry, lipogenesis is the conversion of fatty acids and glycerol into fats, or a metabolic process through which acetyl-CoA is converted to triglyceride for storage in fat. Lipogenesis encompasses both fatty acid and triglyceride synthesis, with the latter being the process by which fatty acids are esterified to glycerol before being packaged into very-low-density lipoprotein (VLDL). Fatty acids are produced in the cytoplasm of cells by repeatedly adding two-carbon units to acetyl-CoA. Triacylglycerol synthesis, on the other hand, occurs in the endoplasmic reticulum membrane of cells by bonding three fatty acid molecules to a glycerol molecule. Both processes take place mainly in liver and adipose tissue. Nevertheless, it also occurs to some extent in other tissues such as the gut and kidney. A review on lipogenesis in the brain was published in 2008 by Lopez and Vidal-Puig. After being packaged into VLDL in the liver, the resulting lipoprotein is then secreted directly into the blood for delivery to peripheral tissues.

Sterol regulatory element-binding proteins (SREBPs) are transcription factors that bind to the sterol regulatory element DNA sequence TCACNCCAC. Mammalian SREBPs are encoded by the genes SREBF1 and SREBF2. SREBPs belong to the basic-helix-loop-helix leucine zipper class of transcription factors. Unactivated SREBPs are attached to the nuclear envelope and endoplasmic reticulum membranes. In cells with low levels of sterols, SREBPs are cleaved to a water-soluble N-terminal domain that is translocated to the nucleus. These activated SREBPs then bind to specific sterol regulatory element DNA sequences, thus upregulating the synthesis of enzymes involved in sterol biosynthesis. Sterols in turn inhibit the cleavage of SREBPs and therefore synthesis of additional sterols is reduced through a negative feed back loop.
In chemistry, de novo synthesis is the synthesis of complex molecules from simple molecules such as sugars or amino acids, as opposed to recycling after partial degradation. For example, nucleotides are not needed in the diet as they can be constructed from small precursor molecules such as formate and aspartate. Methionine, on the other hand, is needed in the diet because while it can be degraded to and then regenerated from homocysteine, it cannot be synthesized de novo.

The liver X receptor (LXR) is a member of the nuclear receptor family of transcription factors and is closely related to nuclear receptors such as the PPARs, FXR and RXR. Liver X receptors (LXRs) are important regulators of cholesterol, fatty acid, and glucose homeostasis. LXRs were earlier classified as orphan nuclear receptors, however, upon discovery of endogenous oxysterols as ligands they were subsequently deorphanized.

Cholesterol 7 alpha-hydroxylase also known as cholesterol 7-alpha-monooxygenase or cytochrome P450 7A1 (CYP7A1) is an enzyme that in humans is encoded by the CYP7A1 gene which has an important role in cholesterol metabolism. It is a cytochrome P450 enzyme, which belongs to the oxidoreductase class, and converts cholesterol to 7-alpha-hydroxycholesterol, the first and rate limiting step in bile acid synthesis.

CREB-binding protein, also known as CREBBP or CBP or KAT3A, is a coactivator encoded by the CREBBP gene in humans, located on chromosome 16p13.3. CBP has intrinsic acetyltransferase functions; it is able to add acetyl groups to both transcription factors as well as histone lysines, the latter of which has been shown to alter chromatin structure making genes more accessible for transcription. This relatively unique acetyltransferase activity is also seen in another transcription enzyme, EP300 (p300). Together, they are known as the p300-CBP coactivator family and are known to associate with more than 16,000 genes in humans; however, while these proteins share many structural features, emerging evidence suggests that these two co-activators may promote transcription of genes with different biological functions.
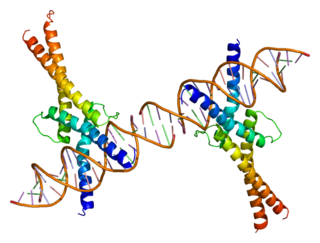
Sterol regulatory element-binding transcription factor 1 (SREBF1) also known as sterol regulatory element-binding protein 1 (SREBP-1) is a protein that in humans is encoded by the SREBF1 gene.

Liver X receptor alpha (LXR-alpha) is a nuclear receptor protein that in humans is encoded by the NR1H3 gene.

Upstream stimulatory factor 1 is a protein that in humans is encoded by the USF1 gene.
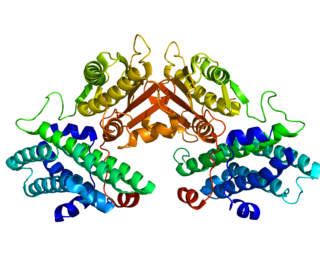
Pyruvate dehydrogenase lipoamide kinase isozyme 4, mitochondrial (PDK4) is an enzyme that in humans is encoded by the PDK4 gene. It codes for an isozyme of pyruvate dehydrogenase kinase.

Insulin induced gene 1, also known as INSIG1, is a protein which in humans is encoded by the INSIG1 gene.

Lipin-1 is a protein that in humans is encoded by the LPIN1 gene.

Max-like protein X is a protein that in humans is encoded by the MLX gene.

Max-interacting transcriptional repressor MAD4 is a protein that in humans is encoded by the MXD4 gene.

MNT is a Max-binding protein that is encoded by the MNT gene

Insulin induced gene 2, also known as INSIG2, is a protein which in humans is encoded by the INSIG2 gene.

Forkhead box protein O1 (FOXO1), also known as forkhead in rhabdomyosarcoma (FKHR), is a protein that in humans is encoded by the FOXO1 gene. FOXO1 is a transcription factor that plays important roles in regulation of gluconeogenesis and glycogenolysis by insulin signaling, and is also central to the decision for a preadipocyte to commit to adipogenesis. It is primarily regulated through phosphorylation on multiple residues; its transcriptional activity is dependent on its phosphorylation state.




















