
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei and in most Archaeal phyla. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30-nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out; however, when wound about histones, this length is reduced to about 90 micrometers (0.09 mm) of 30 nm diameter chromatin fibers.

Histone acetyltransferase p300 also known as p300 HAT or E1A-associated protein p300 also known as EP300 or p300 is an enzyme that, in humans, is encoded by the EP300 gene. It functions as histone acetyltransferase that regulates transcription of genes via chromatin remodeling by allowing histone proteins to wrap DNA less tightly. This enzyme plays an essential role in regulating cell growth and division, prompting cells to mature and assume specialized functions (differentiate), and preventing the growth of cancerous tumors. The p300 protein appears to be critical for normal development before and after birth.
Histone methylation is a process by which methyl groups are transferred to amino acids of histone proteins that make up nucleosomes, which the DNA double helix wraps around to form chromosomes. Methylation of histones can either increase or decrease transcription of genes, depending on which amino acids in the histones are methylated, and how many methyl groups are attached. Methylation events that weaken chemical attractions between histone tails and DNA increase transcription because they enable the DNA to uncoil from nucleosomes so that transcription factor proteins and RNA polymerase can access the DNA. This process is critical for the regulation of gene expression that allows different cells to express different genes.
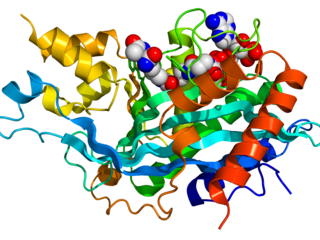
The p300-CBP coactivator family in humans is composed of two closely related transcriptional co-activating proteins :
- p300
- CBP

The autoimmune regulator (AIRE) is a protein that in humans is encoded by the AIRE gene. It is a 13kbp gene on chromosome 21q22.3 that encodes 545 amino acids. AIRE is a transcription factor expressed in the medulla of the thymus. It is part of the mechanism which eliminates self-reactive T cells that would cause autoimmune disease. It exposes T cells to normal, healthy proteins from all parts of the body, and T cells that react to those proteins are destroyed.

Lysine-specific histone demethylase 1A (LSD1) also known as lysine (K)-specific demethylase 1A (KDM1A) is a protein that in humans is encoded by the KDM1A gene. LSD1 is a flavin-dependent monoamine oxidase, which can demethylate mono- and di-methylated lysines, specifically histone 3, lysine 4 (H3K4). Other reported methylated lysine substrates such as histone H3K9 and TP53 have not been biochemically validated. This enzyme plays a critical role in oocyte growth, embryogenesis, hematopoiesis and tissue-specific differentiation. LSD1 was the first histone demethylase to be discovered though more than 30 have since been described.

CREB-binding protein, also known as CREBBP or CBP or KAT3A, is a coactivator encoded by the CREBBP gene in humans, located on chromosome 16p13.3. CBP has intrinsic acetyltransferase functions; it is able to add acetyl groups to both transcription factors as well as histone lysines, the latter of which has been shown to alter chromatin structure making genes more accessible for transcription. This relatively unique acetyltransferase activity is also seen in another transcription enzyme, EP300 (p300). Together, they are known as the p300-CBP coactivator family and are known to associate with more than 16,000 genes in humans; however, while these proteins share many structural features, emerging evidence suggests that these two co-activators may promote transcription of genes with different biological functions.
The nuclear receptor coactivator 2 also known as NCoA-2 is a protein that in humans is encoded by the NCOA2 gene. NCoA-2 is also frequently called glucocorticoid receptor-interacting protein 1 (GRIP1), steroid receptor coactivator-2 (SRC-2), or transcriptional mediators/intermediary factor 2 (TIF2).

Retinoic acid receptor alpha (RAR-α), also known as NR1B1 is a nuclear receptor that in humans is encoded by the RARA gene.

Enhancer of zeste homolog 2 (EZH2) is a histone-lysine N-methyltransferase enzyme encoded by EZH2 gene, that participates in histone methylation and, ultimately, transcriptional repression. EZH2 catalyzes the addition of methyl groups to histone H3 at lysine 27, by using the cofactor S-adenosyl-L-methionine. Methylation activity of EZH2 facilitates heterochromatin formation thereby silences gene function. Remodeling of chromosomal heterochromatin by EZH2 is also required during cell mitosis.

K(lysine) acetyltransferase 6A (KAT6A), is an enzyme that, in humans, is encoded by the KAT6A gene. This gene is located on human chromosome 8, band 8p11.21.

Histone-lysine N-methyltransferase 2D (KMT2D), also known as MLL4 and sometimes MLL2 in humans and Mll4 in mice, is a major mammalian histone H3 lysine 4 (H3K4) mono-methyltransferase. It is part of a family of six Set1-like H3K4 methyltransferases that also contains KMT2A, KMT2B, KMT2C, KMT2F, and KMT2G.

ASH1L is a histone-lysine N-methyltransferase enzyme encoded by the ASH1L gene located at chromosomal band 1q22. ASH1L is the human homolog of Drosophila Ash1.

In biochemistry, the KIX domain (kinase-inducible domain (KID) interacting domain) or CREB binding domain is a protein domain of the eukaryotic transcriptional coactivators CBP and P300. It serves as a docking site for the formation of heterodimers between the coactivator and specific transcription factors. Structurally, the KIX domain is a globular domain consisting of three α-helices and two short 310-helices.
While the cellular and molecular mechanisms of learning and memory have long been a central focus of neuroscience, it is only in recent years that attention has turned to the epigenetic mechanisms behind the dynamic changes in gene transcription responsible for memory formation and maintenance. Epigenetic gene regulation often involves the physical marking of DNA or associated proteins to cause or allow long-lasting changes in gene activity. Epigenetic mechanisms such as DNA methylation and histone modifications have been shown to play an important role in learning and memory.
Cocaine addiction is the compulsive use of cocaine despite adverse consequences. It arises through epigenetic modification and transcriptional regulation of genes in the nucleus accumbens.
Embryonic stem cells are capable of self-renewing and differentiating to the desired fate depending on their position in the body. Stem cell homeostasis is maintained through epigenetic mechanisms that are highly dynamic in regulating the chromatin structure as well as specific gene transcription programs. Epigenetics has been used to refer to changes in gene expression, which are heritable through modifications not affecting the DNA sequence.

Ali Shilatifard is an American biochemist, molecular biologist, the Robert Francis Furchgott Professor and chairman of the department of biochemistry and molecular genetics, and the director of the Simpson Query Institute for Epigenetics at the Northwestern University Feinberg School of Medicine. He has served as a member of the Senior Editorial Board for the journal Science. He also served as the founding Deputy Editor and the first academic Editor for Science's open access journal Science Advances between 2014 and 2023. During his tenure as the editor of Science Advances, the journal brought onboard roughly 50 deputy editors and over 350 associate editors managing over 22,000 annual submissions and roughly 2,000 annual publications, reaching an impact factor of 14.98. He has served on the Scientific Advisory Board (SAB) of Keystone Symposia, Max Planck Society, and Genentech and is a member of the jury for the BBVA Foundation Prize in Medicine.
H3K4me3 is an epigenetic modification to the DNA packaging protein Histone H3 that indicates tri-methylation at the 4th lysine residue of the histone H3 protein and is often involved in the regulation of gene expression. The name denotes the addition of three methyl groups (trimethylation) to the lysine 4 on the histone H3 protein.
In epigenetics, proline isomerization is the effect that cis-trans isomerization of the amino acid proline has on the regulation of gene expression. Similar to aspartic acid, the amino acid proline has the rare property of being able to occupy both cis and trans isomers of its prolyl peptide bonds with ease. Peptidyl-prolyl isomerase, or PPIase, is an enzyme very commonly associated with proline isomerization due to their ability to catalyze the isomerization of prolines. PPIases are present in three types: cyclophilins, FK507-binding proteins, and the parvulins. PPIase enzymes catalyze the transition of proline between cis and trans isomers and are essential to the numerous biological functions controlled and affected by prolyl isomerization Without PPIases, prolyl peptide bonds will slowly switch between cis and trans isomers, a process that can lock proteins in a nonnative structure that can affect render the protein temporarily ineffective. Although this switch can occur on its own, PPIases are responsible for most isomerization of prolyl peptide bonds. The specific amino acid that precedes the prolyl peptide bond also can have an effect on which conformation the bond assumes. For instance, when an aromatic amino acid is bonded to a proline the bond is more favorable to the cis conformation. Cyclophilin A uses an "electrostatic handle" to pull proline into cis and trans formations. Most of these biological functions are affected by the isomerization of proline when one isomer interacts differently than the other, commonly causing an activation/deactivation relationship. As an amino acid, proline is present in many proteins. This aids in the multitude of effects that isomerization of proline can have in different biological mechanisms and functions.























