Proteins
| | This section needs expansion. You can help by adding to it. (January 2015) |
This article is being considered for deletion in accordance with Wikipedia's deletion policy. Please share your thoughts on the matter at this article's deletion discussion page. |
There are many speculations to the number of proteins which could potentially be made, [1] [2] [3] [4] however various sources agree on an estimated 10,000 proteins which are relevant to the human body. [5] [6] [7] [8] These estimates are thanks to recent advances in technology leading to the birth of the field of Proteomics. Another consensus which can also be found is that it takes 20000-25000 Proteins to make a human work and that our cells have the potential to make around a 100 000 different proteins depending on what is most convenient in the environment we live in. [9] [10] Even though this seems like a fair estimate, it is hard to find an emperical database which claims to list more then 10 000 proteins. Another way to approach it is by looking from the bottom up there are ~20,000 protein coding genes in the human genome, (of which it is estimated that 12,733 already have Wikipedia articles (the Gene Wiki) about them). If wone where to Including splice variants, some argue that there could be as many as 500,000 unique human proteins. [11] All of these speculations are based on how the average human at the start of the 21' century looks.
This is a list for only those proteins which have been found in the human body so far. It contains about 1100 unique proteins (all of which have their own wiki article), some of which are listed multiple times waiting to be merged into one row. After the merging work there is still about 9200 proteins yet to be described and listed.
| | This section needs expansion. You can help by adding to it. (January 2015) |
Essential cell biology 5 edition lists: Enzymes(which typically have EC number), Transport proteins(which typically have TC number), Motor Proteins, Storage Proteins, Signal proteins, Receptor Proteins, Transcription Proteins as the most common types of proteins. Finally the make a very broad category of Special-purpose proteins. So far this seems to be the most simple yet sufficient way to split proteins into types
| Column name | Meaning and convention |
|---|---|
| name | The name of the protein in accordance with Gene nomenclature |
| Protein type | Either: Enzymes(EC number should be added here*), Transport proteins(TC number Should be added here), Motor Proteins, Storage Proteins, Signal proteins, Receptor Proteins, Trranscription Proteins or Special-purpose proteins |
| Location | The Tissue, Organs or place and thing in the body where this protein is used |
| Produced by | The cell which typically produce the protein, in case the protein is not produced in the human body please start this column with (Outside the human body). |
| Folding Variations | This refers to the proteins structural domain or Protein domain |
| Produced by the human body | This column indicates weather a protein can be made my the human body. There is a place for proteins in this list which are not made in the human body but which still have a messurable effect on the human body, such as certain types of medicine. |
*An Enzyme Commission number refers to the reaction catalyzed by an enzyme and not the enzyme protein itself. This means that there is often more than one human gene that corresponds to a given EC number.
| | This section needs expansion. You can help by adding to it. (January 2015) |
| DB name | DB website | Provider | Data sources | Revenue/Sponsors sources | Integrates | Wiki article | Desc. | Size | DB type |
|---|---|---|---|---|---|---|---|---|---|
| Database of Macromolecular Movements | http://www.molmovdb.org/ | describes the motions that occur in proteins and other macromolecules, particularly using movies | protein structure databases | ||||||
| Dynameomics | https://ngdc.cncb.ac.cn/databasecommons/database/id/3509 | a data warehouse of molecular dynamics simulations and analyses of proteins representing all known protein fold families | protein structure databases | ||||||
| JenaLib | https://jenalib.leibniz-fli.de/ | the Jena Library of Biological Macromolecules is aimed at a better dissemination of information on three-dimensional biopolymer structures with an emphasis on visualization and analysis. | protein structure databases | ||||||
| OCA | http://oca.weizmann.ac.il/oca-bin/ocamain | KEGG, OMIM, PDBselect, Pfam, PubMed, SCOP, SwissProt, and others | a browser-database for protein structure/function | protein structure databases | |||||
| PDB Lite/SUM | https://www.ebi.ac.uk/pdbe/, http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/, | EMBL | PDB Lite/SUM | lite: derived from OCA, PDB Lite was provided to make it as easy as possible to find and view a macromolecule within the PDB sum: provides an overview macromolecular structures in the PDB, giving schematic diagrams of the molecules in each structure and of the interactions between them | protein structure databases | ||||
| PDBTM | https://web.archive.org/web/20131225065028/http://pdbtm.enzim.hu/ | the Protein Data Bank of Transmembrane Proteins – — a selection of the PDB. | protein structure databases | ||||||
| PDBWiki | a community annotated knowledge base of biological molecular structures | protein structure databases | |||||||
| InterPro | http://www.ebi.ac.uk/interpro/ | ELIXIR infrastructure | European Bioinformatics Institute | EMBL, The Welcome trust, BBSRC | CATH-Gene3D, CDD, HAMAP, MobiDB, PANTHER, Pfam, SMART, SUPERFAMILY, SFLD, TIGRFAMs, | InterPro | classifies proteins into families and predicts the presence of domains and sites | Protein sequence databases | |
| NextProt | https://www.nextprot.org/ | CALIPHO (is a group at the SIB) | Swiss Institute of Bioinformatics | https://www.sib.swiss/about/funding-sources | UniProt, Cellosaurus, Gnomad, IntAct, SRAA Atlas, Uniprot – GOA, BGEE, COSMIC, MassIVE, Peptide atlas | neXtProt | a human protein-centric knowledge resource | Protein sequence databases | |
| Wiki-pi | http://severus.dbmi.pitt.edu/wiki-pi/ | Madhavi K. Ganapathiraju | At present Wiki-Pi contains 48,419 unique interactions among 10,492 proteins. However it is not clear if this is unique proteins[13] | Protein interactoin Database | |||||
| Human Protein Reference Database | Institute of Bioinformatics (IOB), Bangalore, India | Human Protein Reference Database | One source claims 15000 [13] proteins. But it is unclear how many of these are unique | ||||||
| Sanger Institute | Pfam | protein families database of alignments and HMMs | Protein sequence databases | ||||||
| Human Proteinpedia | Institute of Bioinformatics (IOB), Bangalore and Johns Hopkins University, | Human Proteinpedia | The human Proteinpedia is based on HPRD (Human protein reference database)which is a repository hosting over 30,000 human proteins. However it is unclear how many of these are unique proteins | ||||||
| Human Protein Atlas | The Swedish Government | Human Protein Atlas | It contains roughly 10 million IHC images of a bit less then 25,000 antibodies. But once again it is unclear how many of these are unique | ||||||
| Manchester University | PRINTS | a compendium of protein fingerprints | Protein sequence databases | ||||||
| PROSITE | database of protein families and domains | Protein sequence databases | |||||||
| Georgetown University Medical Center [GUMC] | Protein Information Resource | Protein sequence databases | |||||||
| SUPERFAMILY | library of HMMs representing superfamilies and database of (superfamily and family) annotations for all completely sequenced organisms | Protein sequence databases | |||||||
| Swiss Institute of Bioinformatics | Swiss-Prot | protein knowledgebase | Protein sequence databases | ||||||
| NCBI | protein sequence and knowledgebase (National Center for Biotechnology Information) | Protein sequence databases | |||||||
| Protein DataBank in Europe (PDBe), [14] ProteinDatabank in Japan (PDBj), [15] Research Collaboratory for Structural Bioinformatics (RCSB) [16] | Protein Data Bank | (PDB) | Protein structure databases | ||||||
| Structural Classification of Proteins (SCOP) | Protein structure databases | ||||||||
| Protein Structure Classification database | CATH : | Protein structure databases | |||||||
| Sali Lab, UCSF | ModBase | database of comparative protein structure models | Protein model databases | ||||||
| Similarity Matrix of Proteins | SIMAP | database of protein similarities computed using FASTA | Protein model databases | ||||||
| Swiss-model | server and repository for protein structure models | Protein model databases | |||||||
| AAindex | : database of amino acid indices, amino acid mutation matrices, and pair-wise contact potentials | Protein model databases | |||||||
| Samuel Lunenfeld Research Institute | BioGRID | general repository for interaction datasets | Protein-protein and other molecular interactions | ||||||
| RNA-binding protein databas | Protein-protein and other molecular interactions | ||||||||
| Univ. of California | Database of Interacting Proteins | Protein-protein and other molecular interactions | |||||||
| (EMBL-EBI) | IntAct: [17] | open-source database for molecular interactions | Protein-protein and other molecular interactions | ||||||
| String | an open source molecular interaction database to study interactions between proteins | Protein-protein and other molecular interactions | |||||||
| Human Protein Atlas | aims at mapping all the human proteins in cells, tissues and organs | Protein expression databases | |||||||
| ProteinModelPortal | Protein Model Portal of the PSI-Nature Structural Biology Knowledgebase | ?? | ?? | 3D structure protein databases | |||||
| SMR | Database of annotated 3D protein structure models | University of Basel | The Swiss goverment | 3D structure protein databases | |||||
| DisProt | Database of Protein Disorder | ELIXIR infrastructure | Indiana University School of Medicine, Temple University, University of Padua | funding from the European Union’s Horizon 2020 | Swiss Prot/Uni Prot, CATH, Pfam, Europe PMC, BITEM, ECO, Geneontology | DisProt | database of experimental evidences of disorder in proteins | 3D structure protein databases, Protein sequence databases | |
| MobiDB | Database of intrinsically disordered and mobile proteins | John Moult, Christine Orengo, Predrag Radivojac | University of Padua | Italian Goverment | MobiDB | database of intrinsic protein disorder annotation | 3D structure protein databases, Protein sequence databases | ||
| ModBase | Database of Comparative Protein Structure Models | Ursula Pieper, Ben Webb, Narayanan Eswar, Andrej Sali Roberto Sanchez | UCSF, Sali Lab | 3D structure protein databases | |||||
| PDBsum | Pictorial database of 3D structures in the Protein Data Bank | European Bioinformatics Institute 2013 | Wellcome Trust | 3D structure protein databases | |||||
| CCDS | The Consensus CDS protein set database | NCBI | ?? | Sequence databases | |||||
| DDBJ | DNA Data Bank of Japan | ?? | ?? | Sequence databases | |||||
| ENA | European Nucleotide Archive | ?? | ?? | Sequence databases | |||||
| GenBank | GenBank nucleotide sequence database | ?? | ?? | Sequence databases | |||||
| Refseq | NCBI Reference Sequence Database | ?? | ?? | Sequence databases | |||||
| UniGene | Database of computationally identifies transcripts from the same locus | ?? | ?? | Sequence databases | |||||
| UniProtKB | Universal Protein Resource (UniProt) | ?? | ?? | Sequence databases | |||||
| Swiss Prot/Uni Prot | https://www.sib.swiss/swiss-prot,https://www.uniprot.org/ | SIB Swiss Institute of Bioinformatics | European Bioinformatics Institute (EMBL-EBI) | Swiss-Prot has collected over 81 000 variants in roughly 13,000 human protein sequence records from peer-reviewed literature. It is unclear how many unique proteins types are present in the database. |
A restriction enzyme, restriction endonuclease, REase, ENase orrestrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone of the DNA double helix.
A regulatory sequence is a segment of a nucleic acid molecule which is capable of increasing or decreasing the expression of specific genes within an organism. Regulation of gene expression is an essential feature of all living organisms and viruses.

Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two hybridized nucleic acid strands, using energy from ATP hydrolysis. There are many helicases, representing the great variety of processes in which strand separation must be catalyzed. Approximately 1% of eukaryotic genes code for helicases.
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eukaryotes, polyadenylation is part of the process that produces mature mRNA for translation. In many bacteria, the poly(A) tail promotes degradation of the mRNA. It, therefore, forms part of the larger process of gene expression.

Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a duplication event (paralogs), or else a horizontal gene transfer event (xenologs).
In academia, computational immunology is a field of science that encompasses high-throughput genomic and bioinformatics approaches to immunology. The field's main aim is to convert immunological data into computational problems, solve these problems using mathematical and computational approaches and then convert these results into immunologically meaningful interpretations.

KEGG is a collection of databases dealing with genomes, biological pathways, diseases, drugs, and chemical substances. KEGG is utilized for bioinformatics research and education, including data analysis in genomics, metagenomics, metabolomics and other omics studies, modeling and simulation in systems biology, and translational research in drug development.

Amos Bairoch is a Swiss bioinformatician and Professor of Bioinformatics at the Department of Human Protein Sciences of the University of Geneva where he leads the CALIPHO group at the Swiss Institute of Bioinformatics (SIB) combining bioinformatics, curation, and experimental efforts to functionally characterize human proteins.

In molecular biology 7SK is an abundant small nuclear RNA found in metazoans. It plays a role in regulating transcription by controlling the positive transcription elongation factor P-TEFb. 7SK is found in a small nuclear ribonucleoprotein complex (snRNP) with a number of other proteins that regulate the stability and function of the complex.

DNA-(apurinic or apyrimidinic site) lyase is an enzyme that in humans is encoded by the APEX1 gene.

Interferon beta is a protein that in humans is encoded by the IFNB1 gene. The natural and recombinant protein forms have antiviral, antibacterial, and anticancer properties.

Transcription factor SOX-6 is a protein that in humans is encoded by the SOX6 gene.
The Reference Sequence (RefSeq) database is an open access, annotated and curated collection of publicly available nucleotide sequences and their protein products. RefSeq was first introduced in 2000. This database is built by National Center for Biotechnology Information (NCBI), and, unlike GenBank, provides only a single record for each natural biological molecule for major organisms ranging from viruses to bacteria to eukaryotes.

Zinc finger protein 366, also known as DC-SCRIPT, is a protein that in humans is encoded by the ZNF366 gene. The ZNF366 gene was first identified in a DNA comparison study between 85 kb of Fugu rubripes sequence containing 17 genes with its homologous loci in the human draft genome.
Recombinant adeno-associated virus (rAAV) based genome engineering is a genome editing platform centered on the use of recombinant AAV vectors that enables insertion, deletion or substitution of DNA sequences into the genomes of live mammalian cells. The technique builds on Mario Capecchi and Oliver Smithies' Nobel Prize–winning discovery that homologous recombination (HR), a natural hi-fidelity DNA repair mechanism, can be harnessed to perform precise genome alterations in mice. rAAV mediated genome-editing improves the efficiency of this technique to permit genome engineering in any pre-established and differentiated human cell line, which, in contrast to mouse ES cells, have low rates of HR.
Chimeric RNA, sometimes referred to as a fusion transcript, is composed of exons from two or more different genes that have the potential to encode novel proteins. These mRNAs are different from those produced by conventional splicing as they are produced by two or more gene loci.
Model organism databases (MODs) are biological databases, or knowledgebases, dedicated to the provision of in-depth biological data for intensively studied model organisms. MODs allow researchers to easily find background information on large sets of genes, plan experiments efficiently, combine their data with existing knowledge, and construct novel hypotheses. They allow users to analyse results and interpret datasets, and the data they generate are increasingly used to describe less well studied species. Where possible, MODs share common approaches to collect and represent biological information. For example, all MODs use the Gene Ontology (GO) to describe functions, processes and cellular locations of specific gene products. Projects also exist to enable software sharing for curation, visualization and querying between different MODs. Organismal diversity and varying user requirements however mean that MODs are often required to customize capture, display, and provision of data.
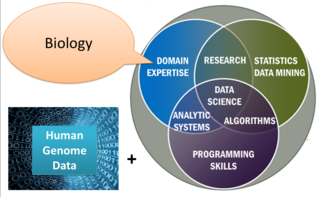
Genome mining describes the exploitation of genomic information for the discovery of biosynthetic pathways of natural products and their possible interactions. It depends on computational technology and bioinformatics tools. The mining process relies on a huge amount of data accessible in genomic databases. By applying data mining algorithms, the data can be used to generate new knowledge in several areas of medicinal chemistry, such as discovering novel natural products.